Vindel: a simple pipeline for checking indel redundancy
Zhiyi Li, Xiaowei Wu, Bin He and Liqing Zhang
Department of Computer Science, Virginia Tech, Blacksburg, VA
Department of Statistics, Virginia Tech, Blacksburg, VA
Abstract:
Background:
With the advance of Next Generation Sequencing (NGS) technologies, a large number
of insertion and deletion variants (indels) have been identified in human populations. Despite
intense effort in variant calling, it has been found that a non-negligible proportion of the identified
indel variants might be false positives and great redundancy exists in the identified indels due to
sequencing errors, artifacts caused by ambiguous alignments, and annotation errors.
Results:
In this paper, we examine indel redundancy in dbSNP, one of the central databases for
indel variants, and develop a standalone computational pipeline, dubbed Vindel, to detect
redundant indels. The pipeline first applies indel position information to form candidate redundant
groups, then performs indel mutations to the reference genome to generate corresponding indel
variant substrings. Finally the indel variant substrings in the same candidate redundant groups are
compared in a pairwise fashion to identify redundant indels. We applied our pipeline to check for
redundancy in dbSNP's human indels. Our pipeline identified approximate 8% redundancy in
insertion type indels, 12% in deletion type indels, and overall 10% for insertions and deletions
combined. These numbers are largely consistent across all human autosomes. We also investigated
indel size distribution and adjacent-indel distance distribution for a better understanding of the
mechanisms generating indel variants.
Conclusions:
Vindel, a simple yet effective computational pipeline, can be used to check whether a
given set of indels are redundant with respect to those already in the database of interest such as
NCBI's dbSNP. Of the ~5.9 million indels we examined, nearly 0.6 million are redundant, revealing a
serious limitation in the current indel annotation. Statistics results prove the pipeline's consistency
on indel redundancy detection for all 22 human chromosomes.
An example of redundant indels |
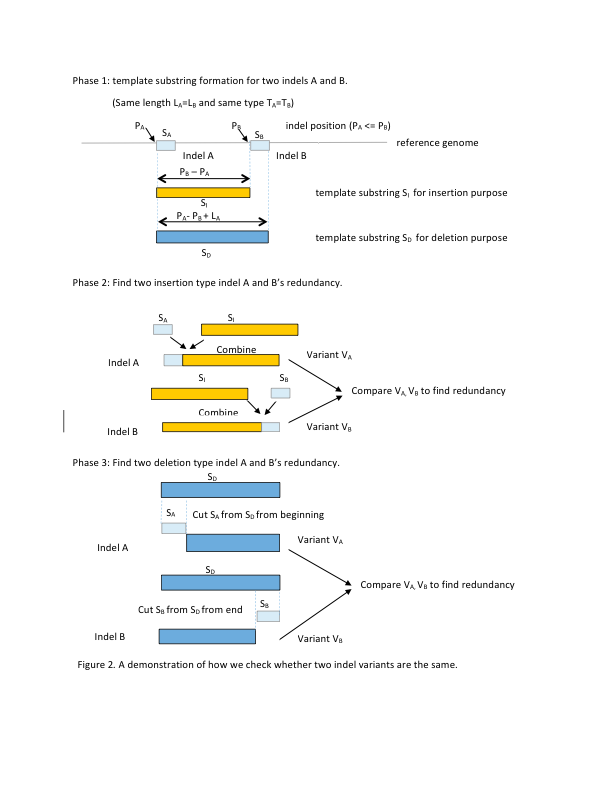
An illustration about how we check redundant indels |
|
|
|
A web tool to check indel redundancy, current can handle basic query for human candidate redundant indels
Source code is available:
Download