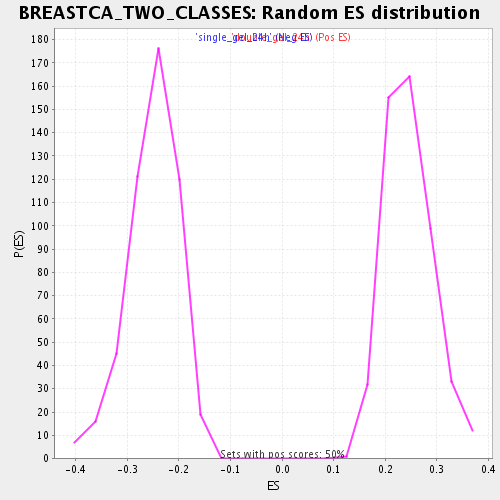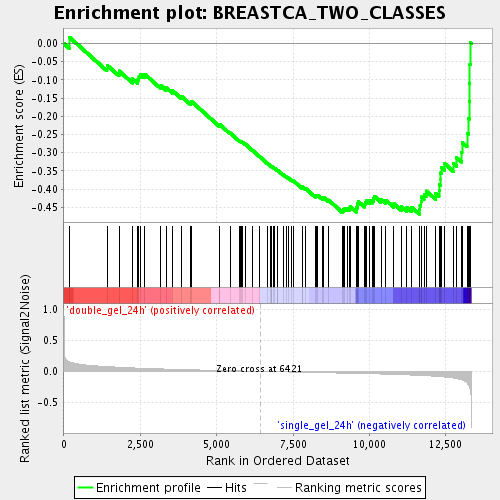
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | single_gel_24h |
| GeneSet | BREASTCA_TWO_CLASSES |
| Enrichment Score (ES) | -0.46894318 |
| Normalized Enrichment Score (NES) | -1.879318 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.10333133 |
| FWER p-Value | 0.816 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | LPIN1 | LPIN1 Entrez, Source | lipin 1 | 182 | 0.151 | 0.0174 | No |
| 2 | MYC | MYC Entrez, Source | v-myc myelocytomatosis viral oncogene homolog (avian) | 1411 | 0.077 | -0.0594 | No |
| 3 | SERPINE1 | SERPINE1 Entrez, Source | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 1805 | 0.067 | -0.0752 | No |
| 4 | FKBP5 | FKBP5 Entrez, Source | FK506 binding protein 5 | 2248 | 0.056 | -0.0970 | No |
| 5 | PARD3 | PARD3 Entrez, Source | par-3 partitioning defective 3 homolog (C. elegans) | 2421 | 0.053 | -0.0990 | No |
| 6 | ERBB2 | ERBB2 Entrez, Source | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | 2456 | 0.053 | -0.0908 | No |
| 7 | CEP57 | CEP57 Entrez, Source | centrosomal protein 57kDa | 2503 | 0.052 | -0.0837 | No |
| 8 | BTG3 | BTG3 Entrez, Source | BTG family, member 3 | 2641 | 0.049 | -0.0839 | No |
| 9 | SLC35B1 | SLC35B1 Entrez, Source | solute carrier family 35, member B1 | 3176 | 0.040 | -0.1159 | No |
| 10 | PHYH | PHYH Entrez, Source | phytanoyl-CoA 2-hydroxylase | 3341 | 0.038 | -0.1206 | No |
| 11 | ZFP36L1 | ZFP36L1 Entrez, Source | zinc finger protein 36, C3H type-like 1 | 3560 | 0.034 | -0.1299 | No |
| 12 | ATN1 | ATN1 Entrez, Source | atrophin 1 | 3850 | 0.030 | -0.1455 | No |
| 13 | GMPS | GMPS Entrez, Source | guanine monphosphate synthetase | 4135 | 0.026 | -0.1615 | No |
| 14 | COG3 | COG3 Entrez, Source | component of oligomeric golgi complex 3 | 4180 | 0.026 | -0.1595 | No |
| 15 | PPP1CB | PPP1CB Entrez, Source | protein phosphatase 1, catalytic subunit, beta isoform | 5079 | 0.015 | -0.2242 | No |
| 16 | FOXM1 | FOXM1 Entrez, Source | forkhead box M1 | 5100 | 0.015 | -0.2226 | No |
| 17 | CDKN2C | CDKN2C Entrez, Source | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | 5438 | 0.011 | -0.2458 | No |
| 18 | SF3A3 | SF3A3 Entrez, Source | splicing factor 3a, subunit 3, 60kDa | 5760 | 0.007 | -0.2685 | No |
| 19 | HADHA | HADHA Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 5790 | 0.007 | -0.2693 | No |
| 20 | UBE2B | UBE2B Entrez, Source | ubiquitin-conjugating enzyme E2B (RAD6 homolog) | 5812 | 0.007 | -0.2695 | No |
| 21 | NUP93 | NUP93 Entrez, Source | nucleoporin 93kDa | 5850 | 0.006 | -0.2711 | No |
| 22 | LRPAP1 | LRPAP1 Entrez, Source | low density lipoprotein receptor-related protein associated protein 1 | 5932 | 0.005 | -0.2761 | No |
| 23 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 6161 | 0.003 | -0.2927 | No |
| 24 | OPRS1 | OPRS1 Entrez, Source | opioid receptor, sigma 1 | 6387 | 0.000 | -0.3096 | No |
| 25 | TUBGCP3 | TUBGCP3 Entrez, Source | tubulin, gamma complex associated protein 3 | 6657 | -0.003 | -0.3294 | No |
| 26 | WWP1 | WWP1 Entrez, Source | WW domain containing E3 ubiquitin protein ligase 1 | 6766 | -0.003 | -0.3369 | No |
| 27 | RAB9 | RAB9 Entrez, Source | RAB9, member RAS oncogene family | 6803 | -0.004 | -0.3388 | No |
| 28 | GRB2 | GRB2 Entrez, Source | growth factor receptor-bound protein 2 | 6805 | -0.004 | -0.3381 | No |
| 29 | MAPK1 | MAPK1 Entrez, Source | mitogen-activated protein kinase 1 | 6842 | -0.004 | -0.3400 | No |
| 30 | GCSH | GCSH Entrez, Source | glycine cleavage system protein H (aminomethyl carrier) | 6899 | -0.005 | -0.3433 | No |
| 31 | TOB1 | TOB1 Entrez, Source | transducer of ERBB2, 1 | 6990 | -0.006 | -0.3489 | No |
| 32 | CD63 | CD63 Entrez, Source | CD63 molecule | 6996 | -0.006 | -0.3481 | No |
| 33 | THOC1 | THOC1 Entrez, Source | THO complex 1 | 7179 | -0.007 | -0.3604 | No |
| 34 | COX6C | COX6C Entrez, Source | cytochrome c oxidase subunit VIc | 7285 | -0.008 | -0.3665 | No |
| 35 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7350 | -0.009 | -0.3695 | No |
| 36 | OAZ1 | OAZ1 Entrez, Source | ornithine decarboxylase antizyme 1 | 7453 | -0.010 | -0.3751 | No |
| 37 | ST13 | ST13 Entrez, Source | suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) | 7511 | -0.011 | -0.3773 | No |
| 38 | PFKP | PFKP Entrez, Source | phosphofructokinase, platelet | 7800 | -0.014 | -0.3962 | No |
| 39 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 7807 | -0.014 | -0.3939 | No |
| 40 | GPX4 | GPX4 Entrez, Source | glutathione peroxidase 4 (phospholipid hydroperoxidase) | 7910 | -0.015 | -0.3986 | No |
| 41 | DDT | DDT Entrez, Source | D-dopachrome tautomerase | 8226 | -0.018 | -0.4186 | No |
| 42 | HDAC3 | HDAC3 Entrez, Source | histone deacetylase 3 | 8258 | -0.018 | -0.4172 | No |
| 43 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 8303 | -0.019 | -0.4167 | No |
| 44 | CTNNB1 | CTNNB1 Entrez, Source | catenin (cadherin-associated protein), beta 1, 88kDa | 8471 | -0.020 | -0.4251 | No |
| 45 | CSDA | CSDA Entrez, Source | cold shock domain protein A | 8491 | -0.021 | -0.4223 | No |
| 46 | GABRP | GABRP Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, pi | 8657 | -0.022 | -0.4301 | No |
| 47 | GDI2 | GDI2 Entrez, Source | GDP dissociation inhibitor 2 | 9107 | -0.027 | -0.4585 | Yes |
| 48 | RDX | RDX Entrez, Source | radixin | 9144 | -0.027 | -0.4555 | Yes |
| 49 | SERBP1 | SERBP1 Entrez, Source | SERPINE1 mRNA binding protein 1 | 9194 | -0.028 | -0.4535 | Yes |
| 50 | GNA12 | GNA12 Entrez, Source | guanine nucleotide binding protein (G protein) alpha 12 | 9281 | -0.029 | -0.4540 | Yes |
| 51 | MTHFD1 | MTHFD1 Entrez, Source | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase | 9334 | -0.029 | -0.4519 | Yes |
| 52 | C10ORF7 | C10ORF7 Entrez, Source | chromosome 10 open reading frame 7 | 9370 | -0.030 | -0.4484 | Yes |
| 53 | CREB1 | CREB1 Entrez, Source | cAMP responsive element binding protein 1 | 9571 | -0.032 | -0.4569 | Yes |
| 54 | DEAF1 | DEAF1 Entrez, Source | deformed epidermal autoregulatory factor 1 (Drosophila) | 9574 | -0.032 | -0.4504 | Yes |
| 55 | FGFR4 | FGFR4 Entrez, Source | fibroblast growth factor receptor 4 | 9601 | -0.033 | -0.4456 | Yes |
| 56 | MNAT1 | MNAT1 Entrez, Source | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 9616 | -0.033 | -0.4399 | Yes |
| 57 | OGT | OGT Entrez, Source | O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) | 9640 | -0.033 | -0.4348 | Yes |
| 58 | DLL1 | DLL1 Entrez, Source | delta-like 1 (Drosophila) | 9837 | -0.036 | -0.4423 | Yes |
| 59 | HP1BP3 | HP1BP3 Entrez, Source | heterochromatin protein 1, binding protein 3 | 9856 | -0.036 | -0.4363 | Yes |
| 60 | THRAP4 | THRAP4 Entrez, Source | thyroid hormone receptor associated protein 4 | 9905 | -0.036 | -0.4325 | Yes |
| 61 | FAH | FAH Entrez, Source | fumarylacetoacetate hydrolase (fumarylacetoacetase) | 9998 | -0.037 | -0.4319 | Yes |
| 62 | CRAT | CRAT Entrez, Source | carnitine acetyltransferase | 10089 | -0.038 | -0.4308 | Yes |
| 63 | SMAD2 | SMAD2 Entrez, Source | SMAD, mothers against DPP homolog 2 (Drosophila) | 10127 | -0.039 | -0.4256 | Yes |
| 64 | NUP155 | NUP155 Entrez, Source | nucleoporin 155kDa | 10149 | -0.039 | -0.4192 | Yes |
| 65 | ATP6AP1 | ATP6AP1 Entrez, Source | ATPase, H+ transporting, lysosomal accessory protein 1 | 10377 | -0.042 | -0.4278 | Yes |
| 66 | EIF2C2 | EIF2C2 Entrez, Source | eukaryotic translation initiation factor 2C, 2 | 10536 | -0.044 | -0.4307 | Yes |
| 67 | POLR2F | POLR2F Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide F | 10800 | -0.048 | -0.4407 | Yes |
| 68 | FABP5 | FABP5 Entrez, Source | fatty acid binding protein 5 (psoriasis-associated) | 11036 | -0.052 | -0.4478 | Yes |
| 69 | MLLT3 | MLLT3 Entrez, Source | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 | 11224 | -0.055 | -0.4505 | Yes |
| 70 | TUBB | TUBB Entrez, Source | tubulin, beta | 11384 | -0.058 | -0.4505 | Yes |
| 71 | CALM2 | CALM2 Entrez, Source | calmodulin 2 (phosphorylase kinase, delta) | 11629 | -0.063 | -0.4560 | Yes |
| 72 | CRYAB | CRYAB Entrez, Source | crystallin, alpha B | 11652 | -0.064 | -0.4445 | Yes |
| 73 | CBX3 | CBX3 Entrez, Source | chromobox homolog 3 (HP1 gamma homolog, Drosophila) | 11686 | -0.064 | -0.4338 | Yes |
| 74 | CDK4 | CDK4 Entrez, Source | cyclin-dependent kinase 4 | 11704 | -0.065 | -0.4217 | Yes |
| 75 | KIF5B | KIF5B Entrez, Source | kinesin family member 5B | 11786 | -0.067 | -0.4141 | Yes |
| 76 | VLDLR | VLDLR Entrez, Source | very low density lipoprotein receptor | 11859 | -0.069 | -0.4054 | Yes |
| 77 | HMGN3 | HMGN3 Entrez, Source | high mobility group nucleosomal binding domain 3 | 12161 | -0.078 | -0.4120 | Yes |
| 78 | ENPP1 | ENPP1 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 1 | 12276 | -0.082 | -0.4038 | Yes |
| 79 | XRCC6 | XRCC6 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 6 (Ku autoantigen, 70kDa) | 12283 | -0.082 | -0.3874 | Yes |
| 80 | PARP1 | PARP1 Entrez, Source | poly (ADP-ribose) polymerase family, member 1 | 12319 | -0.083 | -0.3728 | Yes |
| 81 | PXN | PXN Entrez, Source | paxillin | 12322 | -0.084 | -0.3558 | Yes |
| 82 | GART | GART Entrez, Source | phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase | 12346 | -0.085 | -0.3401 | Yes |
| 83 | GNAS | GNAS Entrez, Source | GNAS complex locus | 12462 | -0.089 | -0.3304 | Yes |
| 84 | MSH2 | MSH2 Entrez, Source | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | 12758 | -0.107 | -0.3305 | Yes |
| 85 | CHEK1 | CHEK1 Entrez, Source | CHK1 checkpoint homolog (S. pombe) | 12847 | -0.115 | -0.3136 | Yes |
| 86 | ALCAM | ALCAM Entrez, Source | activated leukocyte cell adhesion molecule | 13011 | -0.133 | -0.2985 | Yes |
| 87 | PSIP1 | PSIP1 Entrez, Source | PC4 and SFRS1 interacting protein 1 | 13029 | -0.136 | -0.2719 | Yes |
| 88 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 13194 | -0.185 | -0.2462 | Yes |
| 89 | MSN | MSN Entrez, Source | moesin | 13233 | -0.213 | -0.2052 | Yes |
| 90 | IRF1 | IRF1 Entrez, Source | interferon regulatory factor 1 | 13267 | -0.235 | -0.1594 | Yes |
| 91 | FGFR3 | FGFR3 Entrez, Source | fibroblast growth factor receptor 3 (achondroplasia, thanatophoric dwarfism) | 13275 | -0.244 | -0.1097 | Yes |
| 92 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 13284 | -0.263 | -0.0563 | Yes |
| 93 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 13302 | -0.295 | 0.0030 | Yes |