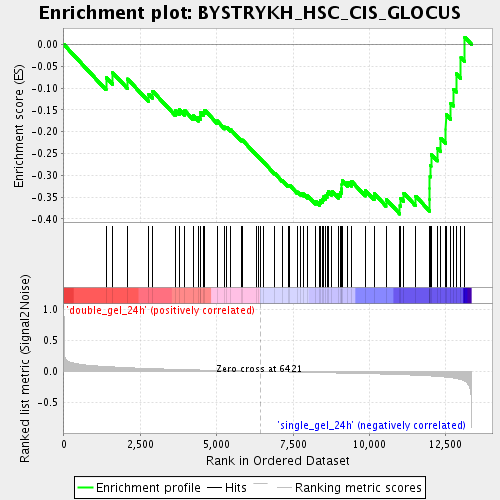
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | single_gel_24h |
| GeneSet | BYSTRYKH_HSC_CIS_GLOCUS |
| Enrichment Score (ES) | -0.3889266 |
| Normalized Enrichment Score (NES) | -1.4827266 |
| Nominal p-value | 0.021097047 |
| FDR q-value | 0.2834038 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GPR56 | GPR56 Entrez, Source | G protein-coupled receptor 56 | 1381 | 0.078 | -0.0752 | No |
| 2 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 1596 | 0.073 | -0.0646 | No |
| 3 | TMEM66 | TMEM66 Entrez, Source | transmembrane protein 66 | 2082 | 0.060 | -0.0788 | No |
| 4 | ST6GALNAC4 | ST6GALNAC4 Entrez, Source | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 | 2775 | 0.047 | -0.1138 | No |
| 5 | GATM | GATM Entrez, Source | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 2899 | 0.045 | -0.1066 | No |
| 6 | GNB4 | GNB4 Entrez, Source | guanine nucleotide binding protein (G protein), beta polypeptide 4 | 3658 | 0.033 | -0.1516 | No |
| 7 | PADI4 | PADI4 Entrez, Source | peptidyl arginine deiminase, type IV | 3776 | 0.031 | -0.1488 | No |
| 8 | TCEB3 | TCEB3 Entrez, Source | transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A) | 3956 | 0.029 | -0.1515 | No |
| 9 | CUTL1 | CUTL1 Entrez, Source | cut-like 1, CCAAT displacement protein (Drosophila) | 4235 | 0.025 | -0.1631 | No |
| 10 | PTPRV | PTPRV Entrez, Source | protein tyrosine phosphatase, receptor type, V (pseudogene) | 4403 | 0.023 | -0.1671 | No |
| 11 | ARNTL | ARNTL Entrez, Source | aryl hydrocarbon receptor nuclear translocator-like | 4468 | 0.022 | -0.1637 | No |
| 12 | SC5DL | SC5DL Entrez, Source | sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, fungal)-like | 4474 | 0.022 | -0.1559 | No |
| 13 | TSPAN8 | TSPAN8 Entrez, Source | tetraspanin 8 | 4578 | 0.021 | -0.1559 | No |
| 14 | NSDHL | NSDHL Entrez, Source | NAD(P) dependent steroid dehydrogenase-like | 4607 | 0.021 | -0.1504 | No |
| 15 | ATP6V1B2 | ATP6V1B2 Entrez, Source | ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 | 5011 | 0.016 | -0.1749 | No |
| 16 | TMCO1 | TMCO1 Entrez, Source | transmembrane and coiled-coil domains 1 | 5262 | 0.013 | -0.1889 | No |
| 17 | CCR2 | CCR2 Entrez, Source | chemokine (C-C motif) receptor 2 | 5326 | 0.012 | -0.1892 | No |
| 18 | DBI | DBI Entrez, Source | diazepam binding inhibitor (GABA receptor modulator, acyl-Coenzyme A binding protein) | 5459 | 0.011 | -0.1952 | No |
| 19 | ARL6IP1 | ARL6IP1 Entrez, Source | ADP-ribosylation factor-like 6 interacting protein 1 | 5810 | 0.007 | -0.2191 | No |
| 20 | ALAD | ALAD Entrez, Source | aminolevulinate, delta-, dehydratase | 5837 | 0.006 | -0.2188 | No |
| 21 | LIG3 | LIG3 Entrez, Source | ligase III, DNA, ATP-dependent | 6312 | 0.001 | -0.2541 | No |
| 22 | LITAF | LITAF Entrez, Source | lipopolysaccharide-induced TNF factor | 6376 | 0.000 | -0.2587 | No |
| 23 | CTSE | CTSE Entrez, Source | cathepsin E | 6446 | -0.000 | -0.2638 | No |
| 24 | ACADL | ACADL Entrez, Source | acyl-Coenzyme A dehydrogenase, long chain | 6542 | -0.001 | -0.2704 | No |
| 25 | PITX2 | PITX2 Entrez, Source | paired-like homeodomain transcription factor 2 | 6901 | -0.005 | -0.2957 | No |
| 26 | PON2 | PON2 Entrez, Source | paraoxonase 2 | 7149 | -0.007 | -0.3117 | No |
| 27 | ATAD1 | ATAD1 Entrez, Source | ATPase family, AAA domain containing 1 | 7341 | -0.009 | -0.3228 | No |
| 28 | RPL26 | RPL26 Entrez, Source | ribosomal protein L26 | 7393 | -0.009 | -0.3232 | No |
| 29 | GLO1 | GLO1 Entrez, Source | glyoxalase I | 7651 | -0.012 | -0.3381 | No |
| 30 | NDNL2 | NDNL2 Entrez, Source | necdin-like 2 | 7749 | -0.013 | -0.3406 | No |
| 31 | GFER | GFER Entrez, Source | growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) | 7843 | -0.014 | -0.3424 | No |
| 32 | AFP | AFP Entrez, Source | alpha-fetoprotein | 7977 | -0.015 | -0.3468 | No |
| 33 | SPG21 | SPG21 Entrez, Source | spastic paraplegia 21, maspardin (autosomal recessive, Mast syndrome) | 8237 | -0.018 | -0.3596 | No |
| 34 | MTIF2 | MTIF2 Entrez, Source | mitochondrial translational initiation factor 2 | 8372 | -0.019 | -0.3625 | No |
| 35 | GATAD2A | GATAD2A Entrez, Source | GATA zinc finger domain containing 2A | 8395 | -0.020 | -0.3569 | No |
| 36 | NFYA | NFYA Entrez, Source | nuclear transcription factor Y, alpha | 8466 | -0.020 | -0.3547 | No |
| 37 | ADPGK | ADPGK Entrez, Source | ADP-dependent glucokinase | 8497 | -0.021 | -0.3494 | No |
| 38 | THUMPD1 | THUMPD1 Entrez, Source | THUMP domain containing 1 | 8574 | -0.022 | -0.3472 | No |
| 39 | DYNLL2 | DYNLL2 Entrez, Source | dynein, light chain, LC8-type 2 | 8613 | -0.022 | -0.3419 | No |
| 40 | TUBB6 | TUBB6 Entrez, Source | tubulin, beta 6 | 8646 | -0.022 | -0.3361 | No |
| 41 | PIGX | PIGX Entrez, Source | phosphatidylinositol glycan anchor biosynthesis, class X | 8772 | -0.023 | -0.3369 | No |
| 42 | CST3 | CST3 Entrez, Source | cystatin C (amyloid angiopathy and cerebral hemorrhage) | 8979 | -0.026 | -0.3429 | No |
| 43 | PSMB6 | PSMB6 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 6 | 9050 | -0.027 | -0.3383 | No |
| 44 | NECAP2 | NECAP2 Entrez, Source | NECAP endocytosis associated 2 | 9087 | -0.027 | -0.3311 | No |
| 45 | FLI1 | FLI1 Entrez, Source | Friend leukemia virus integration 1 | 9098 | -0.027 | -0.3219 | No |
| 46 | IL3RA | IL3RA Entrez, Source | interleukin 3 receptor, alpha (low affinity) | 9102 | -0.027 | -0.3122 | No |
| 47 | CNN3 | CNN3 Entrez, Source | calponin 3, acidic | 9286 | -0.029 | -0.3153 | No |
| 48 | DAP3 | DAP3 Entrez, Source | death associated protein 3 | 9414 | -0.030 | -0.3137 | No |
| 49 | PRDX2 | PRDX2 Entrez, Source | peroxiredoxin 2 | 9864 | -0.036 | -0.3343 | No |
| 50 | HDC | HDC Entrez, Source | histidine decarboxylase | 10155 | -0.039 | -0.3418 | No |
| 51 | GNB1 | GNB1 Entrez, Source | guanine nucleotide binding protein (G protein), beta polypeptide 1 | 10542 | -0.044 | -0.3546 | No |
| 52 | CBR1 | CBR1 Entrez, Source | carbonyl reductase 1 | 10998 | -0.051 | -0.3701 | Yes |
| 53 | MRPS7 | MRPS7 Entrez, Source | mitochondrial ribosomal protein S7 | 11012 | -0.051 | -0.3522 | Yes |
| 54 | AGTRAP | AGTRAP Entrez, Source | angiotensin II receptor-associated protein | 11122 | -0.053 | -0.3408 | Yes |
| 55 | MYOM1 | MYOM1 Entrez, Source | myomesin 1 (skelemin) 185kDa | 11511 | -0.061 | -0.3477 | Yes |
| 56 | PTPRF | PTPRF Entrez, Source | protein tyrosine phosphatase, receptor type, F | 11972 | -0.072 | -0.3558 | Yes |
| 57 | F11R | F11R Entrez, Source | F11 receptor | 11977 | -0.072 | -0.3295 | Yes |
| 58 | MTMR9 | MTMR9 Entrez, Source | myotubularin related protein 9 | 11979 | -0.072 | -0.3030 | Yes |
| 59 | CTSC | CTSC Entrez, Source | cathepsin C | 11993 | -0.073 | -0.2771 | Yes |
| 60 | TFPI | TFPI Entrez, Source | tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) | 12015 | -0.073 | -0.2517 | Yes |
| 61 | DPP7 | DPP7 Entrez, Source | dipeptidyl-peptidase 7 | 12221 | -0.080 | -0.2376 | Yes |
| 62 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 12333 | -0.084 | -0.2148 | Yes |
| 63 | EMP1 | EMP1 Entrez, Source | epithelial membrane protein 1 | 12503 | -0.092 | -0.1938 | Yes |
| 64 | MYO5A | MYO5A Entrez, Source | myosin VA (heavy chain 12, myoxin) | 12504 | -0.092 | -0.1600 | Yes |
| 65 | CSRP1 | CSRP1 Entrez, Source | cysteine and glycine-rich protein 1 | 12660 | -0.100 | -0.1349 | Yes |
| 66 | FLOT1 | FLOT1 Entrez, Source | flotillin 1 | 12744 | -0.106 | -0.1020 | Yes |
| 67 | F2R | F2R Entrez, Source | coagulation factor II (thrombin) receptor | 12839 | -0.114 | -0.0670 | Yes |
| 68 | CRYZ | CRYZ Entrez, Source | crystallin, zeta (quinone reductase) | 12993 | -0.131 | -0.0301 | Yes |
| 69 | RUNX1 | RUNX1 Entrez, Source | runt-related transcription factor 1 (acute myeloid leukemia 1; aml1 oncogene) | 13119 | -0.153 | 0.0168 | Yes |