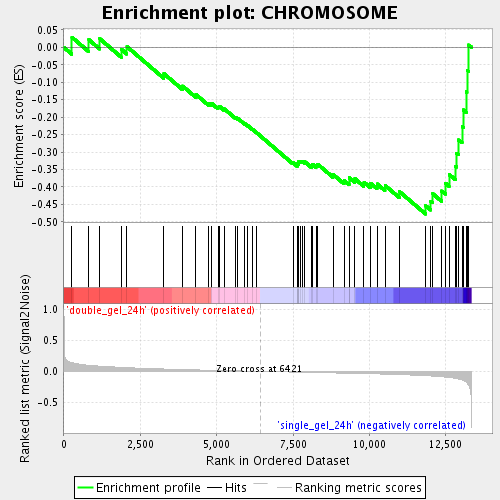
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | single_gel_24h |
| GeneSet | CHROMOSOME |
| Enrichment Score (ES) | -0.47767594 |
| Normalized Enrichment Score (NES) | -1.7310358 |
| Nominal p-value | 0.0018903592 |
| FDR q-value | 0.15103187 |
| FWER p-Value | 0.998 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TTN | TTN Entrez, Source | titin | 262 | 0.140 | 0.0292 | No |
| 2 | JUNB | JUNB Entrez, Source | jun B proto-oncogene | 816 | 0.099 | 0.0221 | No |
| 3 | RGS12 | RGS12 Entrez, Source | regulator of G-protein signalling 12 | 1173 | 0.085 | 0.0249 | No |
| 4 | STAG3 | STAG3 Entrez, Source | stromal antigen 3 | 1880 | 0.065 | -0.0054 | No |
| 5 | SC65 | SC65 Entrez, Source | - | 2063 | 0.061 | 0.0022 | No |
| 6 | UBE2I | UBE2I Entrez, Source | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | 3272 | 0.039 | -0.0751 | No |
| 7 | REPIN1 | REPIN1 Entrez, Source | replication initiator 1 | 3882 | 0.030 | -0.1105 | No |
| 8 | JUND | JUND Entrez, Source | jun D proto-oncogene | 4321 | 0.024 | -0.1350 | No |
| 9 | DNMT3A | DNMT3A Entrez, Source | DNA (cytosine-5-)-methyltransferase 3 alpha | 4742 | 0.019 | -0.1599 | No |
| 10 | JUN | JUN Entrez, Source | jun oncogene | 4841 | 0.018 | -0.1609 | No |
| 11 | NPM2 | NPM2 Entrez, Source | nucleophosmin/nucleoplasmin, 2 | 5044 | 0.016 | -0.1707 | No |
| 12 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 5083 | 0.015 | -0.1683 | No |
| 13 | APTX | APTX Entrez, Source | aprataxin | 5249 | 0.013 | -0.1761 | No |
| 14 | MKI67IP | MKI67IP Entrez, Source | MKI67 (FHA domain) interacting nucleolar phosphoprotein | 5611 | 0.009 | -0.2001 | No |
| 15 | NUFIP1 | NUFIP1 Entrez, Source | nuclear fragile X mental retardation protein interacting protein 1 | 5688 | 0.008 | -0.2031 | No |
| 16 | SUGT1 | SUGT1 Entrez, Source | SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) | 5922 | 0.005 | -0.2187 | No |
| 17 | ZW10 | ZW10 Entrez, Source | ZW10, kinetochore associated, homolog (Drosophila) | 6013 | 0.004 | -0.2240 | No |
| 18 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 6161 | 0.003 | -0.2340 | No |
| 19 | PAFAH1B1 | PAFAH1B1 Entrez, Source | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa | 6305 | 0.001 | -0.2444 | No |
| 20 | KIF22 | KIF22 Entrez, Source | kinesin family member 22 | 7498 | -0.010 | -0.3304 | No |
| 21 | RAN | RAN Entrez, Source | RAN, member RAS oncogene family | 7634 | -0.012 | -0.3364 | No |
| 22 | TOP1 | TOP1 Entrez, Source | topoisomerase (DNA) I | 7635 | -0.012 | -0.3323 | No |
| 23 | RPA2 | RPA2 Entrez, Source | replication protein A2, 32kDa | 7682 | -0.012 | -0.3315 | No |
| 24 | ATRX | ATRX Entrez, Source | alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) | 7684 | -0.012 | -0.3273 | No |
| 25 | HMGN1 | HMGN1 Entrez, Source | high-mobility group nucleosome binding domain 1 | 7731 | -0.013 | -0.3263 | No |
| 26 | XRCC4 | XRCC4 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 4 | 7798 | -0.014 | -0.3265 | No |
| 27 | TERF2IP | TERF2IP Entrez, Source | telomeric repeat binding factor 2, interacting protein | 7871 | -0.014 | -0.3269 | No |
| 28 | TINF2 | TINF2 Entrez, Source | TERF1 (TRF1)-interacting nuclear factor 2 | 8106 | -0.017 | -0.3387 | No |
| 29 | HMGB1 | HMGB1 Entrez, Source | high-mobility group box 1 | 8126 | -0.017 | -0.3342 | No |
| 30 | RFC1 | RFC1 Entrez, Source | replication factor C (activator 1) 1, 145kDa | 8252 | -0.018 | -0.3373 | No |
| 31 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 8303 | -0.019 | -0.3345 | No |
| 32 | BUB3 | BUB3 Entrez, Source | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) | 8807 | -0.024 | -0.3639 | No |
| 33 | TNP1 | TNP1 Entrez, Source | transition protein 1 (during histone to protamine replacement) | 9171 | -0.028 | -0.3816 | No |
| 34 | SMC1A | SMC1A Entrez, Source | structural maintenance of chromosomes 1A | 9336 | -0.029 | -0.3836 | No |
| 35 | TOP2A | TOP2A Entrez, Source | topoisomerase (DNA) II alpha 170kDa | 9337 | -0.029 | -0.3733 | No |
| 36 | LRPPRC | LRPPRC Entrez, Source | leucine-rich PPR-motif containing | 9523 | -0.032 | -0.3762 | No |
| 37 | H2AFY | H2AFY Entrez, Source | H2A histone family, member Y | 9820 | -0.035 | -0.3861 | No |
| 38 | KLHDC3 | KLHDC3 Entrez, Source | kelch domain containing 3 | 10035 | -0.038 | -0.3891 | No |
| 39 | RFC2 | RFC2 Entrez, Source | replication factor C (activator 1) 2, 40kDa | 10249 | -0.040 | -0.3912 | No |
| 40 | DCTN2 | DCTN2 Entrez, Source | dynactin 2 (p50) | 10509 | -0.044 | -0.3955 | No |
| 41 | ZWINT | ZWINT Entrez, Source | ZW10 interactor | 10978 | -0.051 | -0.4130 | No |
| 42 | PAM | PAM Entrez, Source | peptidylglycine alpha-amidating monooxygenase | 11839 | -0.068 | -0.4538 | Yes |
| 43 | PSEN1 | PSEN1 Entrez, Source | presenilin 1 (Alzheimer disease 3) | 12004 | -0.073 | -0.4407 | Yes |
| 44 | APC | APC Entrez, Source | adenomatosis polyposis coli | 12071 | -0.075 | -0.4194 | Yes |
| 45 | BUB1B | BUB1B Entrez, Source | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | 12364 | -0.086 | -0.4114 | Yes |
| 46 | TUBG1 | TUBG1 Entrez, Source | tubulin, gamma 1 | 12486 | -0.090 | -0.3889 | Yes |
| 47 | RB1 | RB1 Entrez, Source | retinoblastoma 1 (including osteosarcoma) | 12607 | -0.097 | -0.3640 | Yes |
| 48 | TMPO | TMPO Entrez, Source | thymopoietin | 12812 | -0.111 | -0.3405 | Yes |
| 49 | CHEK1 | CHEK1 Entrez, Source | CHK1 checkpoint homolog (S. pombe) | 12847 | -0.115 | -0.3030 | Yes |
| 50 | RFC3 | RFC3 Entrez, Source | replication factor C (activator 1) 3, 38kDa | 12915 | -0.123 | -0.2651 | Yes |
| 51 | PSEN2 | PSEN2 Entrez, Source | presenilin 2 (Alzheimer disease 4) | 13032 | -0.136 | -0.2262 | Yes |
| 52 | MAF | MAF Entrez, Source | v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) | 13072 | -0.143 | -0.1790 | Yes |
| 53 | CLASP1 | CLASP1 Entrez, Source | cytoplasmic linker associated protein 1 | 13168 | -0.170 | -0.1269 | Yes |
| 54 | TIMELESS | TIMELESS Entrez, Source | timeless homolog (Drosophila) | 13192 | -0.182 | -0.0652 | Yes |
| 55 | SMC4 | SMC4 Entrez, Source | structural maintenance of chromosomes 4 | 13248 | -0.219 | 0.0071 | Yes |

