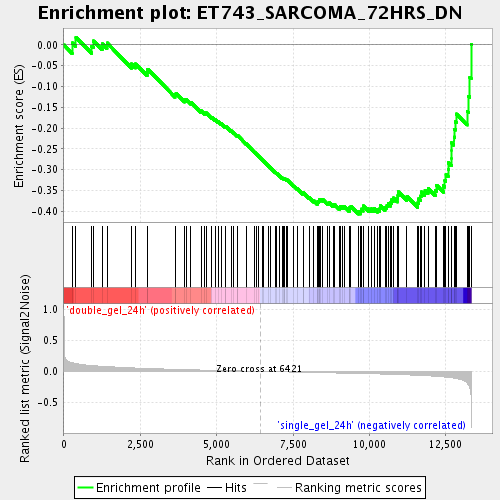
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | single_gel_24h |
| GeneSet | ET743_SARCOMA_72HRS_DN |
| Enrichment Score (ES) | -0.40690133 |
| Normalized Enrichment Score (NES) | -1.6867269 |
| Nominal p-value | 0.001996008 |
| FDR q-value | 0.16579705 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | WNT5B | WNT5B Entrez, Source | wingless-type MMTV integration site family, member 5B | 270 | 0.139 | 0.0048 | No |
| 2 | HSPH1 | HSPH1 Entrez, Source | heat shock 105kDa/110kDa protein 1 | 393 | 0.126 | 0.0184 | No |
| 3 | ACSL3 | ACSL3 Entrez, Source | acyl-CoA synthetase long-chain family member 3 | 906 | 0.095 | -0.0031 | No |
| 4 | CPT1A | CPT1A Entrez, Source | carnitine palmitoyltransferase 1A (liver) | 963 | 0.093 | 0.0096 | No |
| 5 | XBP1 | XBP1 Entrez, Source | X-box binding protein 1 | 1253 | 0.082 | 0.0026 | No |
| 6 | MYC | MYC Entrez, Source | v-myc myelocytomatosis viral oncogene homolog (avian) | 1411 | 0.077 | 0.0047 | No |
| 7 | CDH11 | CDH11 Entrez, Source | cadherin 11, type 2, OB-cadherin (osteoblast) | 2216 | 0.057 | -0.0457 | No |
| 8 | STK17B | STK17B Entrez, Source | serine/threonine kinase 17b (apoptosis-inducing) | 2348 | 0.055 | -0.0457 | No |
| 9 | RAB5A | RAB5A Entrez, Source | RAB5A, member RAS oncogene family | 2725 | 0.048 | -0.0655 | No |
| 10 | GNL2 | GNL2 Entrez, Source | guanine nucleotide binding protein-like 2 (nucleolar) | 2742 | 0.047 | -0.0582 | No |
| 11 | JAG1 | JAG1 Entrez, Source | jagged 1 (Alagille syndrome) | 3636 | 0.033 | -0.1196 | No |
| 12 | CCNH | CCNH Entrez, Source | cyclin H | 3667 | 0.033 | -0.1160 | No |
| 13 | UBE2D2 | UBE2D2 Entrez, Source | ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | 3952 | 0.029 | -0.1321 | No |
| 14 | CLCN7 | CLCN7 Entrez, Source | chloride channel 7 | 4002 | 0.028 | -0.1307 | No |
| 15 | ELOVL6 | ELOVL6 Entrez, Source | ELOVL family member 6, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast) | 4158 | 0.026 | -0.1377 | No |
| 16 | PJA2 | PJA2 Entrez, Source | praja 2, RING-H2 motif containing | 4488 | 0.022 | -0.1585 | No |
| 17 | TXNRD1 | TXNRD1 Entrez, Source | thioredoxin reductase 1 | 4604 | 0.021 | -0.1635 | No |
| 18 | BCL2L1 | BCL2L1 Entrez, Source | BCL2-like 1 | 4653 | 0.020 | -0.1635 | No |
| 19 | PRKCBP1 | PRKCBP1 Entrez, Source | protein kinase C binding protein 1 | 4831 | 0.018 | -0.1736 | No |
| 20 | TARDBP | TARDBP Entrez, Source | TAR DNA binding protein | 4950 | 0.017 | -0.1795 | No |
| 21 | TGIF | TGIF Entrez, Source | TGFB-induced factor (TALE family homeobox) | 5050 | 0.015 | -0.1842 | No |
| 22 | RABGGTB | RABGGTB Entrez, Source | Rab geranylgeranyltransferase, beta subunit | 5146 | 0.014 | -0.1888 | No |
| 23 | BIRC2 | BIRC2 Entrez, Source | baculoviral IAP repeat-containing 2 | 5286 | 0.013 | -0.1970 | No |
| 24 | HSPA8 | HSPA8 Entrez, Source | heat shock 70kDa protein 8 | 5301 | 0.013 | -0.1957 | No |
| 25 | ARFGAP1 | ARFGAP1 Entrez, Source | ADP-ribosylation factor GTPase activating protein 1 | 5472 | 0.010 | -0.2067 | No |
| 26 | RAB1A | RAB1A Entrez, Source | RAB1A, member RAS oncogene family | 5561 | 0.009 | -0.2116 | No |
| 27 | EIF3S10 | EIF3S10 Entrez, Source | eukaryotic translation initiation factor 3, subunit 10 theta, 150/170kDa | 5672 | 0.008 | -0.2185 | No |
| 28 | NUFIP1 | NUFIP1 Entrez, Source | nuclear fragile X mental retardation protein interacting protein 1 | 5688 | 0.008 | -0.2181 | No |
| 29 | SMAD4 | SMAD4 Entrez, Source | SMAD, mothers against DPP homolog 4 (Drosophila) | 5961 | 0.005 | -0.2378 | No |
| 30 | SLC2A1 | SLC2A1 Entrez, Source | solute carrier family 2 (facilitated glucose transporter), member 1 | 6242 | 0.002 | -0.2586 | No |
| 31 | FNTA | FNTA Entrez, Source | farnesyltransferase, CAAX box, alpha | 6311 | 0.001 | -0.2635 | No |
| 32 | ACTL6A | ACTL6A Entrez, Source | actin-like 6A | 6365 | 0.001 | -0.2674 | No |
| 33 | DNAJA2 | DNAJA2 Entrez, Source | DnaJ (Hsp40) homolog, subfamily A, member 2 | 6495 | -0.001 | -0.2771 | No |
| 34 | RBM17 | RBM17 Entrez, Source | RNA binding motif protein 17 | 6541 | -0.001 | -0.2802 | No |
| 35 | MAPK14 | MAPK14 Entrez, Source | mitogen-activated protein kinase 14 | 6688 | -0.003 | -0.2907 | No |
| 36 | PLAG1 | PLAG1 Entrez, Source | pleiomorphic adenoma gene 1 | 6754 | -0.003 | -0.2950 | No |
| 37 | DDX20 | DDX20 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 | 6922 | -0.005 | -0.3068 | No |
| 38 | CDC5L | CDC5L Entrez, Source | CDC5 cell division cycle 5-like (S. pombe) | 6966 | -0.005 | -0.3091 | No |
| 39 | RAB2 | RAB2 Entrez, Source | RAB2, member RAS oncogene family | 7067 | -0.006 | -0.3155 | No |
| 40 | ATP2A2 | ATP2A2 Entrez, Source | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | 7144 | -0.007 | -0.3200 | No |
| 41 | UBE2D3 | UBE2D3 Entrez, Source | ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) | 7172 | -0.007 | -0.3207 | No |
| 42 | PSMD12 | PSMD12 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 | 7220 | -0.008 | -0.3229 | No |
| 43 | CCT4 | CCT4 Entrez, Source | chaperonin containing TCP1, subunit 4 (delta) | 7230 | -0.008 | -0.3221 | No |
| 44 | PSMA5 | PSMA5 Entrez, Source | proteasome (prosome, macropain) subunit, alpha type, 5 | 7274 | -0.008 | -0.3238 | No |
| 45 | HDAC2 | HDAC2 Entrez, Source | histone deacetylase 2 | 7306 | -0.009 | -0.3246 | No |
| 46 | EIF4B | EIF4B Entrez, Source | eukaryotic translation initiation factor 4B | 7529 | -0.011 | -0.3394 | No |
| 47 | TOP1 | TOP1 Entrez, Source | topoisomerase (DNA) I | 7635 | -0.012 | -0.3452 | No |
| 48 | FBXL3 | FBXL3 Entrez, Source | F-box and leucine-rich repeat protein 3 | 7824 | -0.014 | -0.3569 | No |
| 49 | SON | SON Entrez, Source | SON DNA binding protein | 7838 | -0.014 | -0.3554 | No |
| 50 | HSPE1 | HSPE1 Entrez, Source | heat shock 10kDa protein 1 (chaperonin 10) | 8023 | -0.016 | -0.3664 | No |
| 51 | THRAP3 | THRAP3 Entrez, Source | thyroid hormone receptor associated protein 3 | 8178 | -0.017 | -0.3749 | No |
| 52 | GTF2B | GTF2B Entrez, Source | general transcription factor IIB | 8289 | -0.019 | -0.3799 | No |
| 53 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 8303 | -0.019 | -0.3775 | No |
| 54 | FKBP1A | FKBP1A Entrez, Source | FK506 binding protein 1A, 12kDa | 8328 | -0.019 | -0.3758 | No |
| 55 | SNIP1 | SNIP1 Entrez, Source | Smad nuclear interacting protein 1 | 8354 | -0.019 | -0.3742 | No |
| 56 | MTIF2 | MTIF2 Entrez, Source | mitochondrial translational initiation factor 2 | 8372 | -0.019 | -0.3720 | No |
| 57 | PSMC6 | PSMC6 Entrez, Source | proteasome (prosome, macropain) 26S subunit, ATPase, 6 | 8411 | -0.020 | -0.3713 | No |
| 58 | CXXC5 | CXXC5 Entrez, Source | CXXC finger 5 | 8472 | -0.020 | -0.3721 | No |
| 59 | PPP2CB | PPP2CB Entrez, Source | protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform | 8641 | -0.022 | -0.3808 | No |
| 60 | PSMA4 | PSMA4 Entrez, Source | proteasome (prosome, macropain) subunit, alpha type, 4 | 8680 | -0.023 | -0.3795 | No |
| 61 | BUB3 | BUB3 Entrez, Source | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) | 8807 | -0.024 | -0.3847 | No |
| 62 | FOSL1 | FOSL1 Entrez, Source | FOS-like antigen 1 | 8863 | -0.025 | -0.3844 | No |
| 63 | PPP2R1B | PPP2R1B Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), beta isoform | 9012 | -0.026 | -0.3909 | No |
| 64 | ERCC3 | ERCC3 Entrez, Source | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | 9037 | -0.026 | -0.3879 | No |
| 65 | GDI2 | GDI2 Entrez, Source | GDP dissociation inhibitor 2 | 9107 | -0.027 | -0.3882 | No |
| 66 | MID1IP1 | MID1IP1 Entrez, Source | MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) | 9178 | -0.028 | -0.3884 | No |
| 67 | TOP2A | TOP2A Entrez, Source | topoisomerase (DNA) II alpha 170kDa | 9337 | -0.029 | -0.3950 | No |
| 68 | PSMB1 | PSMB1 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 1 | 9355 | -0.030 | -0.3910 | No |
| 69 | EXOC2 | EXOC2 Entrez, Source | exocyst complex component 2 | 9392 | -0.030 | -0.3882 | No |
| 70 | OGT | OGT Entrez, Source | O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) | 9640 | -0.033 | -0.4009 | Yes |
| 71 | PRIM2A | PRIM2A Entrez, Source | primase, polypeptide 2A, 58kDa | 9718 | -0.034 | -0.4005 | Yes |
| 72 | RNF14 | RNF14 Entrez, Source | ring finger protein 14 | 9724 | -0.034 | -0.3946 | Yes |
| 73 | ARFGEF2 | ARFGEF2 Entrez, Source | ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) | 9790 | -0.035 | -0.3932 | Yes |
| 74 | ARHGEF2 | ARHGEF2 Entrez, Source | rho/rac guanine nucleotide exchange factor (GEF) 2 | 9794 | -0.035 | -0.3871 | Yes |
| 75 | SPIN | SPIN Entrez, Source | spindlin | 9979 | -0.037 | -0.3943 | Yes |
| 76 | TDG | TDG Entrez, Source | thymine-DNA glycosylase | 10070 | -0.038 | -0.3942 | Yes |
| 77 | CDK7 | CDK7 Entrez, Source | cyclin-dependent kinase 7 (MO15 homolog, Xenopus laevis, cdk-activating kinase) | 10156 | -0.039 | -0.3936 | Yes |
| 78 | XRCC5 | XRCC5 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining; Ku autoantigen, 80kDa) | 10271 | -0.040 | -0.3949 | Yes |
| 79 | PANX1 | PANX1 Entrez, Source | pannexin 1 | 10341 | -0.041 | -0.3927 | Yes |
| 80 | PPP1CC | PPP1CC Entrez, Source | protein phosphatase 1, catalytic subunit, gamma isoform | 10356 | -0.041 | -0.3862 | Yes |
| 81 | GGPS1 | GGPS1 Entrez, Source | geranylgeranyl diphosphate synthase 1 | 10510 | -0.044 | -0.3899 | Yes |
| 82 | NCOA6 | NCOA6 Entrez, Source | nuclear receptor coactivator 6 | 10571 | -0.044 | -0.3864 | Yes |
| 83 | GCNT1 | GCNT1 Entrez, Source | glucosaminyl (N-acetyl) transferase 1, core 2 (beta-1,6-N-acetylglucosaminyltransferase) | 10607 | -0.045 | -0.3809 | Yes |
| 84 | EPS15 | EPS15 Entrez, Source | epidermal growth factor receptor pathway substrate 15 | 10698 | -0.046 | -0.3793 | Yes |
| 85 | CDC23 | CDC23 Entrez, Source | CDC23 (cell division cycle 23, yeast, homolog) | 10723 | -0.047 | -0.3727 | Yes |
| 86 | HERC4 | HERC4 Entrez, Source | hect domain and RLD 4 | 10776 | -0.048 | -0.3680 | Yes |
| 87 | RAP1B | RAP1B Entrez, Source | RAP1B, member of RAS oncogene family | 10907 | -0.050 | -0.3688 | Yes |
| 88 | CRK | CRK Entrez, Source | v-crk sarcoma virus CT10 oncogene homolog (avian) | 10930 | -0.050 | -0.3614 | Yes |
| 89 | CRSP6 | CRSP6 Entrez, Source | cofactor required for Sp1 transcriptional activation, subunit 6, 77kDa | 10942 | -0.050 | -0.3531 | Yes |
| 90 | MCTS1 | MCTS1 Entrez, Source | malignant T cell amplified sequence 1 | 11227 | -0.056 | -0.3645 | Yes |
| 91 | LBR | LBR Entrez, Source | lamin B receptor | 11583 | -0.062 | -0.3801 | Yes |
| 92 | CDC27 | CDC27 Entrez, Source | cell division cycle 27 | 11606 | -0.063 | -0.3704 | Yes |
| 93 | SUPV3L1 | SUPV3L1 Entrez, Source | suppressor of var1, 3-like 1 (S. cerevisiae) | 11668 | -0.064 | -0.3634 | Yes |
| 94 | USP3 | USP3 Entrez, Source | ubiquitin specific peptidase 3 | 11693 | -0.065 | -0.3535 | Yes |
| 95 | SFRS9 | SFRS9 Entrez, Source | splicing factor, arginine/serine-rich 9 | 11816 | -0.068 | -0.3504 | Yes |
| 96 | CFLAR | CFLAR Entrez, Source | CASP8 and FADD-like apoptosis regulator | 11917 | -0.070 | -0.3453 | Yes |
| 97 | FADD | FADD Entrez, Source | Fas (TNFRSF6)-associated via death domain | 12156 | -0.078 | -0.3491 | Yes |
| 98 | CYHR1 | CYHR1 Entrez, Source | cysteine/histidine-rich 1 | 12194 | -0.079 | -0.3376 | Yes |
| 99 | VEGFC | VEGFC Entrez, Source | vascular endothelial growth factor C | 12416 | -0.088 | -0.3384 | Yes |
| 100 | GIPC1 | GIPC1 Entrez, Source | GIPC PDZ domain containing family, member 1 | 12467 | -0.090 | -0.3259 | Yes |
| 101 | EMP1 | EMP1 Entrez, Source | epithelial membrane protein 1 | 12503 | -0.092 | -0.3119 | Yes |
| 102 | LYN | LYN Entrez, Source | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | 12577 | -0.095 | -0.3002 | Yes |
| 103 | SIRPA | SIRPA Entrez, Source | signal-regulatory protein alpha | 12585 | -0.096 | -0.2833 | Yes |
| 104 | MAT2A | MAT2A Entrez, Source | methionine adenosyltransferase II, alpha | 12685 | -0.102 | -0.2723 | Yes |
| 105 | FZD1 | FZD1 Entrez, Source | frizzled homolog 1 (Drosophila) | 12689 | -0.102 | -0.2540 | Yes |
| 106 | POLD3 | POLD3 Entrez, Source | polymerase (DNA-directed), delta 3, accessory subunit | 12692 | -0.103 | -0.2356 | Yes |
| 107 | PIK3C3 | PIK3C3 Entrez, Source | phosphoinositide-3-kinase, class 3 | 12766 | -0.108 | -0.2214 | Yes |
| 108 | WEE1 | WEE1 Entrez, Source | WEE1 homolog (S. pombe) | 12787 | -0.110 | -0.2031 | Yes |
| 109 | TMPO | TMPO Entrez, Source | thymopoietin | 12812 | -0.111 | -0.1847 | Yes |
| 110 | FAS | FAS Entrez, Source | Fas (TNF receptor superfamily, member 6) | 12841 | -0.114 | -0.1661 | Yes |
| 111 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 13194 | -0.185 | -0.1592 | Yes |
| 112 | SPRY2 | SPRY2 Entrez, Source | sprouty homolog 2 (Drosophila) | 13247 | -0.219 | -0.1234 | Yes |
| 113 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 13284 | -0.263 | -0.0785 | Yes |
| 114 | IL1RL1 | IL1RL1 Entrez, Source | interleukin 1 receptor-like 1 | 13329 | -0.457 | 0.0010 | Yes |