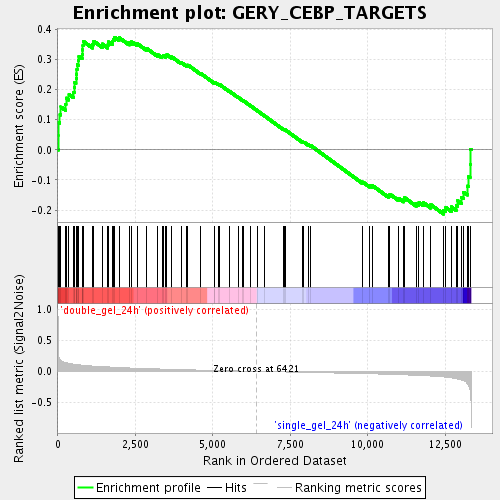
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | GERY_CEBP_TARGETS |
| Enrichment Score (ES) | 0.37401146 |
| Normalized Enrichment Score (NES) | 1.4763596 |
| Nominal p-value | 0.028625954 |
| FDR q-value | 0.2811185 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FOS | FOS Entrez, Source | v-fos FBJ murine osteosarcoma viral oncogene homolog | 11 | 0.331 | 0.0489 | Yes |
| 2 | ZFAND2A | ZFAND2A Entrez, Source | zinc finger, AN1-type domain 2A | 15 | 0.290 | 0.0922 | Yes |
| 3 | DNAJB9 | DNAJB9 Entrez, Source | DnaJ (Hsp40) homolog, subfamily B, member 9 | 66 | 0.194 | 0.1176 | Yes |
| 4 | NR1D1 | NR1D1 Entrez, Source | nuclear receptor subfamily 1, group D, member 1 | 96 | 0.180 | 0.1424 | Yes |
| 5 | HMGCR | HMGCR Entrez, Source | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | 250 | 0.141 | 0.1521 | Yes |
| 6 | IFRD1 | IFRD1 Entrez, Source | interferon-related developmental regulator 1 | 263 | 0.140 | 0.1722 | Yes |
| 7 | DDIT3 | DDIT3 Entrez, Source | DNA-damage-inducible transcript 3 | 356 | 0.129 | 0.1846 | Yes |
| 8 | ATF3 | ATF3 Entrez, Source | activating transcription factor 3 | 502 | 0.118 | 0.1913 | Yes |
| 9 | IER2 | IER2 Entrez, Source | immediate early response 2 | 528 | 0.115 | 0.2067 | Yes |
| 10 | TNFAIP6 | TNFAIP6 Entrez, Source | tumor necrosis factor, alpha-induced protein 6 | 543 | 0.114 | 0.2228 | Yes |
| 11 | RGS16 | RGS16 Entrez, Source | regulator of G-protein signalling 16 | 584 | 0.111 | 0.2365 | Yes |
| 12 | MAFK | MAFK Entrez, Source | v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian) | 599 | 0.110 | 0.2520 | Yes |
| 13 | PPARG | PPARG Entrez, Source | peroxisome proliferative activated receptor, gamma | 612 | 0.109 | 0.2675 | Yes |
| 14 | MYD116 | 623 | 0.109 | 0.2831 | Yes | ||
| 15 | MVD | MVD Entrez, Source | mevalonate (diphospho) decarboxylase | 658 | 0.107 | 0.2965 | Yes |
| 16 | BTG2 | BTG2 Entrez, Source | BTG family, member 2 | 677 | 0.105 | 0.3110 | Yes |
| 17 | BAG3 | BAG3 Entrez, Source | BCL2-associated athanogene 3 | 783 | 0.100 | 0.3181 | Yes |
| 18 | LSS | LSS Entrez, Source | lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) | 797 | 0.100 | 0.3321 | Yes |
| 19 | HMOX1 | HMOX1 Entrez, Source | heme oxygenase (decycling) 1 | 806 | 0.099 | 0.3464 | Yes |
| 20 | JUNB | JUNB Entrez, Source | jun B proto-oncogene | 816 | 0.099 | 0.3606 | Yes |
| 21 | LPL | LPL Entrez, Source | lipoprotein lipase | 1116 | 0.087 | 0.3510 | Yes |
| 22 | S100A3 | S100A3 Entrez, Source | S100 calcium binding protein A3 | 1154 | 0.085 | 0.3611 | Yes |
| 23 | GADD45B | GADD45B Entrez, Source | growth arrest and DNA-damage-inducible, beta | 1431 | 0.077 | 0.3518 | Yes |
| 24 | SERPINI1 | SERPINI1 Entrez, Source | serpin peptidase inhibitor, clade I (neuroserpin), member 1 | 1615 | 0.072 | 0.3488 | Yes |
| 25 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 1628 | 0.072 | 0.3587 | Yes |
| 26 | ACSL1 | ACSL1 Entrez, Source | acyl-CoA synthetase long-chain family member 1 | 1761 | 0.068 | 0.3590 | Yes |
| 27 | FGF7 | FGF7 Entrez, Source | fibroblast growth factor 7 (keratinocyte growth factor) | 1778 | 0.068 | 0.3680 | Yes |
| 28 | ABCA1 | ABCA1 Entrez, Source | ATP-binding cassette, sub-family A (ABC1), member 1 | 1832 | 0.067 | 0.3740 | Yes |
| 29 | TRIB3 | TRIB3 Entrez, Source | tribbles homolog 3 (Drosophila) | 1974 | 0.063 | 0.3729 | No |
| 30 | CXCL5 | CXCL5 Entrez, Source | chemokine (C-X-C motif) ligand 5 | 2302 | 0.055 | 0.3565 | No |
| 31 | DCN | DCN Entrez, Source | decorin | 2379 | 0.054 | 0.3589 | No |
| 32 | SH3GL3 | SH3GL3 Entrez, Source | SH3-domain GRB2-like 3 | 2557 | 0.051 | 0.3531 | No |
| 33 | IGF2 | IGF2 Entrez, Source | insulin-like growth factor 2 (somatomedin A) | 2858 | 0.045 | 0.3373 | No |
| 34 | ANGPTL4 | ANGPTL4 Entrez, Source | angiopoietin-like 4 | 3226 | 0.040 | 0.3156 | No |
| 35 | PIM1 | PIM1 Entrez, Source | pim-1 oncogene | 3360 | 0.037 | 0.3112 | No |
| 36 | EREG | EREG Entrez, Source | epiregulin | 3391 | 0.037 | 0.3144 | No |
| 37 | GCH1 | GCH1 Entrez, Source | GTP cyclohydrolase 1 (dopa-responsive dystonia) | 3481 | 0.035 | 0.3130 | No |
| 38 | HGF | HGF Entrez, Source | hepatocyte growth factor (hepapoietin A; scatter factor) | 3517 | 0.035 | 0.3156 | No |
| 39 | CD68 | CD68 Entrez, Source | CD68 molecule | 3671 | 0.033 | 0.3090 | No |
| 40 | XDH | XDH Entrez, Source | xanthine dehydrogenase | 3980 | 0.029 | 0.2901 | No |
| 41 | ELOVL6 | ELOVL6 Entrez, Source | ELOVL family member 6, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast) | 4158 | 0.026 | 0.2807 | No |
| 42 | HP | HP Entrez, Source | haptoglobin | 4195 | 0.026 | 0.2819 | No |
| 43 | NSDHL | NSDHL Entrez, Source | NAD(P) dependent steroid dehydrogenase-like | 4607 | 0.021 | 0.2540 | No |
| 44 | NRP1 | NRP1 Entrez, Source | neuropilin 1 | 5047 | 0.015 | 0.2232 | No |
| 45 | CSPG2 | CSPG2 Entrez, Source | chondroitin sulfate proteoglycan 2 (versican) | 5062 | 0.015 | 0.2244 | No |
| 46 | FOXG1 | 5170 | 0.014 | 0.2185 | No | ||
| 47 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 5228 | 0.013 | 0.2162 | No |
| 48 | ORM1 | ORM1 Entrez, Source | orosomucoid 1 | 5539 | 0.010 | 0.1943 | No |
| 49 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 5819 | 0.007 | 0.1742 | No |
| 50 | C3 | C3 Entrez, Source | complement component 3 | 5962 | 0.005 | 0.1643 | No |
| 51 | LCN2 | LCN2 Entrez, Source | lipocalin 2 (oncogene 24p3) | 5994 | 0.005 | 0.1626 | No |
| 52 | HBEGF | HBEGF Entrez, Source | heparin-binding EGF-like growth factor | 6200 | 0.002 | 0.1475 | No |
| 53 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 6435 | -0.000 | 0.1299 | No |
| 54 | MKNK2 | MKNK2 Entrez, Source | MAP kinase interacting serine/threonine kinase 2 | 6661 | -0.003 | 0.1133 | No |
| 55 | CD1D1 | 7269 | -0.008 | 0.0688 | No | ||
| 56 | APBB1IP | APBB1IP Entrez, Source | amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein | 7296 | -0.009 | 0.0681 | No |
| 57 | DUSP1 | DUSP1 Entrez, Source | dual specificity phosphatase 1 | 7336 | -0.009 | 0.0665 | No |
| 58 | ACTA2 | ACTA2 Entrez, Source | actin, alpha 2, smooth muscle, aorta | 7897 | -0.014 | 0.0264 | No |
| 59 | RNH1 | RNH1 Entrez, Source | ribonuclease/angiogenin inhibitor 1 | 7938 | -0.015 | 0.0257 | No |
| 60 | RDH11 | RDH11 Entrez, Source | retinol dehydrogenase 11 (all-trans/9-cis/11-cis) | 8080 | -0.017 | 0.0175 | No |
| 61 | ABCD2 | ABCD2 Entrez, Source | ATP-binding cassette, sub-family D (ALD), member 2 | 8162 | -0.017 | 0.0140 | No |
| 62 | BAIAP2 | BAIAP2 Entrez, Source | BAI1-associated protein 2 | 9835 | -0.035 | -0.1068 | No |
| 63 | PLAUR | PLAUR Entrez, Source | plasminogen activator, urokinase receptor | 10063 | -0.038 | -0.1182 | No |
| 64 | RGS2 | RGS2 Entrez, Source | regulator of G-protein signalling 2, 24kDa | 10143 | -0.039 | -0.1183 | No |
| 65 | CLU | CLU Entrez, Source | clusterin | 10667 | -0.046 | -0.1509 | No |
| 66 | NR2F6 | NR2F6 Entrez, Source | nuclear receptor subfamily 2, group F, member 6 | 10711 | -0.046 | -0.1472 | No |
| 67 | RASL11B | RASL11B Entrez, Source | RAS-like, family 11, member B | 11001 | -0.051 | -0.1613 | No |
| 68 | SERPINB2 | SERPINB2 Entrez, Source | serpin peptidase inhibitor, clade B (ovalbumin), member 2 | 11162 | -0.054 | -0.1653 | No |
| 69 | CRABP2 | CRABP2 Entrez, Source | cellular retinoic acid binding protein 2 | 11193 | -0.055 | -0.1593 | No |
| 70 | S100A8 | S100A8 Entrez, Source | S100 calcium binding protein A8 | 11562 | -0.062 | -0.1778 | No |
| 71 | CRYAB | CRYAB Entrez, Source | crystallin, alpha B | 11652 | -0.064 | -0.1750 | No |
| 72 | PEA15 | PEA15 Entrez, Source | phosphoprotein enriched in astrocytes 15 | 11797 | -0.067 | -0.1757 | No |
| 73 | RRAD | RRAD Entrez, Source | Ras-related associated with diabetes | 12036 | -0.074 | -0.1826 | No |
| 74 | PRNP | PRNP Entrez, Source | prion protein (p27-30) (Creutzfeldt-Jakob disease, Gerstmann-Strausler-Scheinker syndrome, fatal familial insomnia) | 12448 | -0.089 | -0.2003 | No |
| 75 | CD47 | CD47 Entrez, Source | CD47 molecule | 12518 | -0.093 | -0.1915 | No |
| 76 | PER2 | PER2 Entrez, Source | period homolog 2 (Drosophila) | 12697 | -0.103 | -0.1895 | No |
| 77 | KLF4 | KLF4 Entrez, Source | Kruppel-like factor 4 (gut) | 12854 | -0.115 | -0.1840 | No |
| 78 | CXCL1 | CXCL1 Entrez, Source | chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | 12901 | -0.121 | -0.1692 | No |
| 79 | DBP | DBP Entrez, Source | D site of albumin promoter (albumin D-box) binding protein | 13022 | -0.135 | -0.1580 | No |
| 80 | ID2 | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 13084 | -0.146 | -0.1406 | No |
| 81 | PLAT | PLAT Entrez, Source | plasminogen activator, tissue | 13214 | -0.201 | -0.1202 | No |
| 82 | FHL2 | FHL2 Entrez, Source | four and a half LIM domains 2 | 13253 | -0.222 | -0.0898 | No |
| 83 | PROCR | PROCR Entrez, Source | protein C receptor, endothelial (EPCR) | 13305 | -0.301 | -0.0484 | No |
| 84 | ID3 | ID3 Entrez, Source | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | 13314 | -0.341 | 0.0021 | No |

