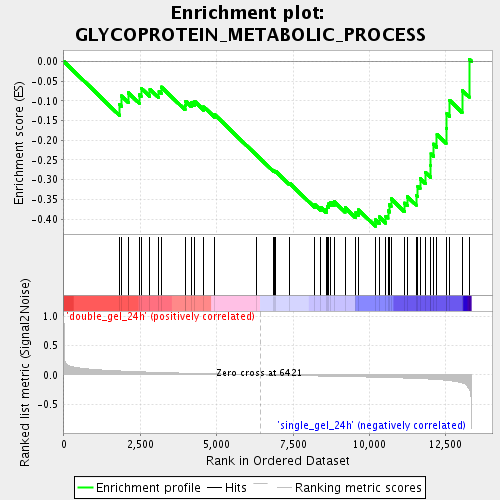
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | single_gel_24h |
| GeneSet | GLYCOPROTEIN_METABOLIC_PROCESS |
| Enrichment Score (ES) | -0.4181506 |
| Normalized Enrichment Score (NES) | -1.5015877 |
| Nominal p-value | 0.031007752 |
| FDR q-value | 0.2746658 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ST8SIA3 | ST8SIA3 Entrez, Source | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 | 1820 | 0.067 | -0.1092 | No |
| 2 | SERP1 | SERP1 Entrez, Source | - | 1887 | 0.065 | -0.0872 | No |
| 3 | BACE2 | BACE2 Entrez, Source | beta-site APP-cleaving enzyme 2 | 2107 | 0.060 | -0.0790 | No |
| 4 | GALNT7 | GALNT7 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) | 2468 | 0.052 | -0.0845 | No |
| 5 | ST8SIA2 | ST8SIA2 Entrez, Source | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 | 2528 | 0.051 | -0.0678 | No |
| 6 | LRP2 | LRP2 Entrez, Source | low density lipoprotein-related protein 2 | 2815 | 0.046 | -0.0703 | No |
| 7 | LDLR | LDLR Entrez, Source | low density lipoprotein receptor (familial hypercholesterolemia) | 3110 | 0.041 | -0.0753 | No |
| 8 | CD37 | CD37 Entrez, Source | CD37 molecule | 3190 | 0.040 | -0.0646 | No |
| 9 | FUT2 | FUT2 Entrez, Source | fucosyltransferase 2 (secretor status included) | 3966 | 0.029 | -0.1109 | No |
| 10 | BACE1 | BACE1 Entrez, Source | beta-site APP-cleaving enzyme 1 | 3984 | 0.029 | -0.1004 | No |
| 11 | COG3 | COG3 Entrez, Source | component of oligomeric golgi complex 3 | 4180 | 0.026 | -0.1043 | No |
| 12 | FUT4 | FUT4 Entrez, Source | fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) | 4282 | 0.025 | -0.1016 | No |
| 13 | TSPAN8 | TSPAN8 Entrez, Source | tetraspanin 8 | 4578 | 0.021 | -0.1151 | No |
| 14 | CLN3 | CLN3 Entrez, Source | ceroid-lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) | 4935 | 0.017 | -0.1350 | No |
| 15 | ALG2 | ALG2 Entrez, Source | asparagine-linked glycosylation 2 homolog (S. cerevisiae, alpha-1,3-mannosyltransferase) | 6291 | 0.001 | -0.2363 | No |
| 16 | PSENEN | PSENEN Entrez, Source | presenilin enhancer 2 homolog (C. elegans) | 6855 | -0.004 | -0.2769 | No |
| 17 | ST8SIA4 | ST8SIA4 Entrez, Source | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 | 6893 | -0.005 | -0.2778 | No |
| 18 | GALNT1 | GALNT1 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) | 6935 | -0.005 | -0.2789 | No |
| 19 | APOA4 | APOA4 Entrez, Source | apolipoprotein A-IV | 7392 | -0.009 | -0.3093 | No |
| 20 | RPN1 | RPN1 Entrez, Source | ribophorin I | 8214 | -0.018 | -0.3636 | No |
| 21 | POMGNT1 | POMGNT1 Entrez, Source | protein O-linked mannose beta1,2-N-acetylglucosaminyltransferase | 8405 | -0.020 | -0.3698 | No |
| 22 | TXNDC4 | TXNDC4 Entrez, Source | thioredoxin domain containing 4 (endoplasmic reticulum) | 8591 | -0.022 | -0.3747 | No |
| 23 | MGEA5 | MGEA5 Entrez, Source | meningioma expressed antigen 5 (hyaluronidase) | 8610 | -0.022 | -0.3670 | No |
| 24 | ALK | ALK Entrez, Source | anaplastic lymphoma kinase (Ki-1) | 8660 | -0.022 | -0.3614 | No |
| 25 | ABCG1 | ABCG1 Entrez, Source | ATP-binding cassette, sub-family G (WHITE), member 1 | 8730 | -0.023 | -0.3571 | No |
| 26 | MAN2B1 | MAN2B1 Entrez, Source | mannosidase, alpha, class 2B, member 1 | 8845 | -0.024 | -0.3555 | No |
| 27 | MPDU1 | MPDU1 Entrez, Source | mannose-P-dolichol utilization defect 1 | 9201 | -0.028 | -0.3706 | No |
| 28 | B3GALT4 | B3GALT4 Entrez, Source | UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 | 9555 | -0.032 | -0.3839 | No |
| 29 | OGT | OGT Entrez, Source | O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) | 9640 | -0.033 | -0.3765 | No |
| 30 | NCSTN | NCSTN Entrez, Source | nicastrin | 10195 | -0.039 | -0.4018 | Yes |
| 31 | POFUT1 | POFUT1 Entrez, Source | protein O-fucosyltransferase 1 | 10320 | -0.041 | -0.3943 | Yes |
| 32 | MGAT4A | MGAT4A Entrez, Source | mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A | 10535 | -0.044 | -0.3922 | Yes |
| 33 | GCNT1 | GCNT1 Entrez, Source | glucosaminyl (N-acetyl) transferase 1, core 2 (beta-1,6-N-acetylglucosaminyltransferase) | 10607 | -0.045 | -0.3790 | Yes |
| 34 | LIPC | LIPC Entrez, Source | lipase, hepatic | 10652 | -0.046 | -0.3635 | Yes |
| 35 | GCS1 | GCS1 Entrez, Source | - | 10708 | -0.046 | -0.3484 | Yes |
| 36 | FUT9 | FUT9 Entrez, Source | fucosyltransferase 9 (alpha (1,3) fucosyltransferase) | 11148 | -0.054 | -0.3592 | Yes |
| 37 | ALG5 | ALG5 Entrez, Source | asparagine-linked glycosylation 5 homolog (S. cerevisiae, dolichyl-phosphate beta-glucosyltransferase) | 11240 | -0.056 | -0.3430 | Yes |
| 38 | DPM2 | DPM2 Entrez, Source | dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit | 11529 | -0.061 | -0.3394 | Yes |
| 39 | FUT1 | FUT1 Entrez, Source | fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group) | 11582 | -0.062 | -0.3177 | Yes |
| 40 | FUT8 | FUT8 Entrez, Source | fucosyltransferase 8 (alpha (1,6) fucosyltransferase) | 11656 | -0.064 | -0.2967 | Yes |
| 41 | ALG8 | ALG8 Entrez, Source | asparagine-linked glycosylation 8 homolog (S. cerevisiae, alpha-1,3-glucosyltransferase) | 11838 | -0.068 | -0.2821 | Yes |
| 42 | PSEN1 | PSEN1 Entrez, Source | presenilin 1 (Alzheimer disease 3) | 12004 | -0.073 | -0.2643 | Yes |
| 43 | COG7 | COG7 Entrez, Source | component of oligomeric golgi complex 7 | 12012 | -0.073 | -0.2346 | Yes |
| 44 | AGA | AGA Entrez, Source | aspartylglucosaminidase | 12094 | -0.076 | -0.2094 | Yes |
| 45 | APH1A | APH1A Entrez, Source | anterior pharynx defective 1 homolog A (C. elegans) | 12209 | -0.080 | -0.1850 | Yes |
| 46 | GCNT3 | GCNT3 Entrez, Source | glucosaminyl (N-acetyl) transferase 3, mucin type | 12507 | -0.092 | -0.1694 | Yes |
| 47 | POMT1 | POMT1 Entrez, Source | protein-O-mannosyltransferase 1 | 12533 | -0.093 | -0.1327 | Yes |
| 48 | B3GALNT1 | B3GALNT1 Entrez, Source | beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) | 12625 | -0.098 | -0.0990 | Yes |
| 49 | PSEN2 | PSEN2 Entrez, Source | presenilin 2 (Alzheimer disease 4) | 13032 | -0.136 | -0.0731 | Yes |
| 50 | TM4SF4 | TM4SF4 Entrez, Source | transmembrane 4 L six family member 4 | 13263 | -0.233 | 0.0059 | Yes |