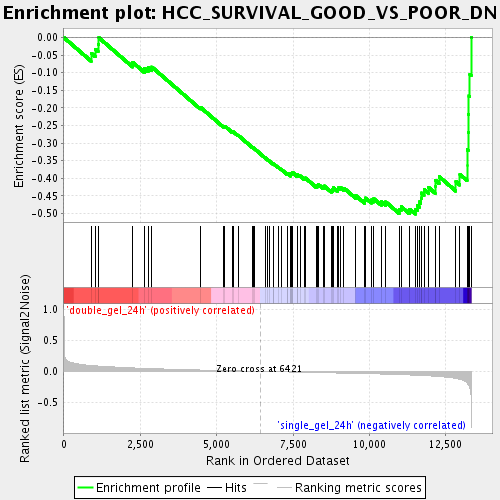
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | single_gel_24h |
| GeneSet | HCC_SURVIVAL_GOOD_VS_POOR_DN |
| Enrichment Score (ES) | -0.50210375 |
| Normalized Enrichment Score (NES) | -1.9550062 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.06362642 |
| FWER p-Value | 0.497 |

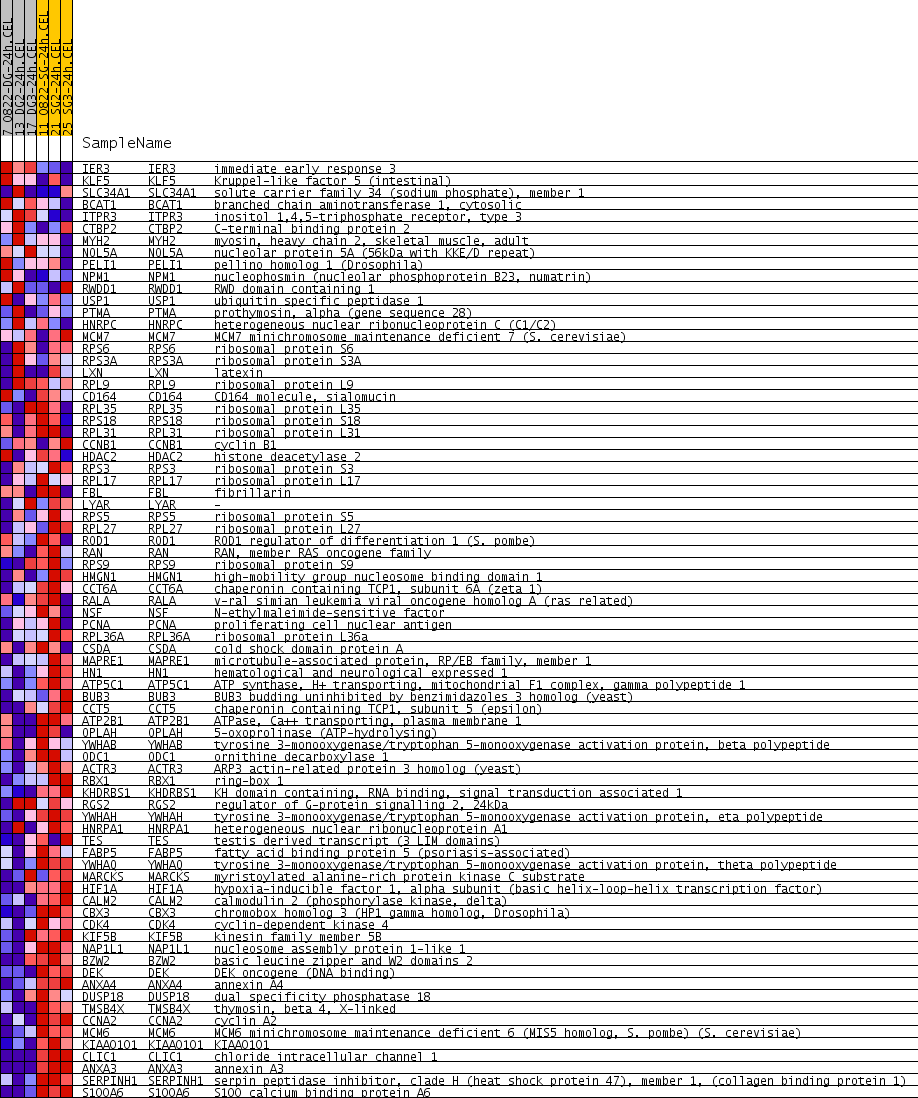
| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IER3 | IER3 Entrez, Source | immediate early response 3 | 890 | 0.096 | -0.0443 | No |
| 2 | KLF5 | KLF5 Entrez, Source | Kruppel-like factor 5 (intestinal) | 1027 | 0.091 | -0.0329 | No |
| 3 | SLC34A1 | SLC34A1 Entrez, Source | solute carrier family 34 (sodium phosphate), member 1 | 1120 | 0.087 | -0.0191 | No |
| 4 | BCAT1 | BCAT1 Entrez, Source | branched chain aminotransferase 1, cytosolic | 1144 | 0.086 | -0.0004 | No |
| 5 | ITPR3 | ITPR3 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 3 | 2251 | 0.056 | -0.0704 | No |
| 6 | CTBP2 | CTBP2 Entrez, Source | C-terminal binding protein 2 | 2635 | 0.049 | -0.0875 | No |
| 7 | MYH2 | MYH2 Entrez, Source | myosin, heavy chain 2, skeletal muscle, adult | 2757 | 0.047 | -0.0854 | No |
| 8 | NOL5A | NOL5A Entrez, Source | nucleolar protein 5A (56kDa with KKE/D repeat) | 2877 | 0.045 | -0.0837 | No |
| 9 | PELI1 | PELI1 Entrez, Source | pellino homolog 1 (Drosophila) | 4464 | 0.022 | -0.1979 | No |
| 10 | NPM1 | NPM1 Entrez, Source | nucleophosmin (nucleolar phosphoprotein B23, numatrin) | 5222 | 0.013 | -0.2517 | No |
| 11 | RWDD1 | RWDD1 Entrez, Source | RWD domain containing 1 | 5268 | 0.013 | -0.2520 | No |
| 12 | USP1 | USP1 Entrez, Source | ubiquitin specific peptidase 1 | 5517 | 0.010 | -0.2683 | No |
| 13 | PTMA | PTMA Entrez, Source | prothymosin, alpha (gene sequence 28) | 5545 | 0.010 | -0.2681 | No |
| 14 | HNRPC | HNRPC Entrez, Source | heterogeneous nuclear ribonucleoprotein C (C1/C2) | 5697 | 0.008 | -0.2775 | No |
| 15 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 6161 | 0.003 | -0.3118 | No |
| 16 | RPS6 | RPS6 Entrez, Source | ribosomal protein S6 | 6196 | 0.002 | -0.3137 | No |
| 17 | RPS3A | RPS3A Entrez, Source | ribosomal protein S3A | 6224 | 0.002 | -0.3153 | No |
| 18 | LXN | LXN Entrez, Source | latexin | 6581 | -0.002 | -0.3417 | No |
| 19 | RPL9 | RPL9 Entrez, Source | ribosomal protein L9 | 6671 | -0.003 | -0.3478 | No |
| 20 | CD164 | CD164 Entrez, Source | CD164 molecule, sialomucin | 6728 | -0.003 | -0.3513 | No |
| 21 | RPL35 | RPL35 Entrez, Source | ribosomal protein L35 | 6847 | -0.004 | -0.3592 | No |
| 22 | RPS18 | RPS18 Entrez, Source | ribosomal protein S18 | 6873 | -0.004 | -0.3600 | No |
| 23 | RPL31 | RPL31 Entrez, Source | ribosomal protein L31 | 7012 | -0.006 | -0.3690 | No |
| 24 | CCNB1 | CCNB1 Entrez, Source | cyclin B1 | 7114 | -0.007 | -0.3750 | No |
| 25 | HDAC2 | HDAC2 Entrez, Source | histone deacetylase 2 | 7306 | -0.009 | -0.3874 | No |
| 26 | RPS3 | RPS3 Entrez, Source | ribosomal protein S3 | 7308 | -0.009 | -0.3854 | No |
| 27 | RPL17 | RPL17 Entrez, Source | ribosomal protein L17 | 7424 | -0.010 | -0.3917 | No |
| 28 | FBL | FBL Entrez, Source | fibrillarin | 7429 | -0.010 | -0.3897 | No |
| 29 | LYAR | LYAR Entrez, Source | - | 7434 | -0.010 | -0.3876 | No |
| 30 | RPS5 | RPS5 Entrez, Source | ribosomal protein S5 | 7442 | -0.010 | -0.3857 | No |
| 31 | RPL27 | RPL27 Entrez, Source | ribosomal protein L27 | 7475 | -0.010 | -0.3857 | No |
| 32 | ROD1 | ROD1 Entrez, Source | ROD1 regulator of differentiation 1 (S. pombe) | 7487 | -0.010 | -0.3841 | No |
| 33 | RAN | RAN Entrez, Source | RAN, member RAS oncogene family | 7634 | -0.012 | -0.3923 | No |
| 34 | RPS9 | RPS9 Entrez, Source | ribosomal protein S9 | 7639 | -0.012 | -0.3897 | No |
| 35 | HMGN1 | HMGN1 Entrez, Source | high-mobility group nucleosome binding domain 1 | 7731 | -0.013 | -0.3935 | No |
| 36 | CCT6A | CCT6A Entrez, Source | chaperonin containing TCP1, subunit 6A (zeta 1) | 7874 | -0.014 | -0.4008 | No |
| 37 | RALA | RALA Entrez, Source | v-ral simian leukemia viral oncogene homolog A (ras related) | 7900 | -0.015 | -0.3992 | No |
| 38 | NSF | NSF Entrez, Source | N-ethylmaleimide-sensitive factor | 8257 | -0.018 | -0.4217 | No |
| 39 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 8303 | -0.019 | -0.4206 | No |
| 40 | RPL36A | RPL36A Entrez, Source | ribosomal protein L36a | 8327 | -0.019 | -0.4178 | No |
| 41 | CSDA | CSDA Entrez, Source | cold shock domain protein A | 8491 | -0.021 | -0.4252 | No |
| 42 | MAPRE1 | MAPRE1 Entrez, Source | microtubule-associated protein, RP/EB family, member 1 | 8516 | -0.021 | -0.4220 | No |
| 43 | HN1 | HN1 Entrez, Source | hematological and neurological expressed 1 | 8753 | -0.023 | -0.4342 | No |
| 44 | ATP5C1 | ATP5C1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | 8795 | -0.024 | -0.4316 | No |
| 45 | BUB3 | BUB3 Entrez, Source | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) | 8807 | -0.024 | -0.4267 | No |
| 46 | CCT5 | CCT5 Entrez, Source | chaperonin containing TCP1, subunit 5 (epsilon) | 8949 | -0.025 | -0.4313 | No |
| 47 | ATP2B1 | ATP2B1 Entrez, Source | ATPase, Ca++ transporting, plasma membrane 1 | 8980 | -0.026 | -0.4274 | No |
| 48 | OPLAH | OPLAH Entrez, Source | 5-oxoprolinase (ATP-hydrolysing) | 9043 | -0.027 | -0.4257 | No |
| 49 | YWHAB | YWHAB Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | 9164 | -0.028 | -0.4282 | No |
| 50 | ODC1 | ODC1 Entrez, Source | ornithine decarboxylase 1 | 9554 | -0.032 | -0.4498 | No |
| 51 | ACTR3 | ACTR3 Entrez, Source | ARP3 actin-related protein 3 homolog (yeast) | 9840 | -0.036 | -0.4628 | No |
| 52 | RBX1 | RBX1 Entrez, Source | ring-box 1 | 9866 | -0.036 | -0.4562 | No |
| 53 | KHDRBS1 | KHDRBS1 Entrez, Source | KH domain containing, RNA binding, signal transduction associated 1 | 10061 | -0.038 | -0.4618 | No |
| 54 | RGS2 | RGS2 Entrez, Source | regulator of G-protein signalling 2, 24kDa | 10143 | -0.039 | -0.4586 | No |
| 55 | YWHAH | YWHAH Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | 10384 | -0.042 | -0.4668 | No |
| 56 | HNRPA1 | HNRPA1 Entrez, Source | heterogeneous nuclear ribonucleoprotein A1 | 10533 | -0.044 | -0.4674 | No |
| 57 | TES | TES Entrez, Source | testis derived transcript (3 LIM domains) | 10972 | -0.051 | -0.4884 | Yes |
| 58 | FABP5 | FABP5 Entrez, Source | fatty acid binding protein 5 (psoriasis-associated) | 11036 | -0.052 | -0.4808 | Yes |
| 59 | YWHAQ | YWHAQ Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | 11312 | -0.057 | -0.4879 | Yes |
| 60 | MARCKS | MARCKS Entrez, Source | myristoylated alanine-rich protein kinase C substrate | 11501 | -0.060 | -0.4877 | Yes |
| 61 | HIF1A | HIF1A Entrez, Source | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | 11568 | -0.062 | -0.4779 | Yes |
| 62 | CALM2 | CALM2 Entrez, Source | calmodulin 2 (phosphorylase kinase, delta) | 11629 | -0.063 | -0.4674 | Yes |
| 63 | CBX3 | CBX3 Entrez, Source | chromobox homolog 3 (HP1 gamma homolog, Drosophila) | 11686 | -0.064 | -0.4562 | Yes |
| 64 | CDK4 | CDK4 Entrez, Source | cyclin-dependent kinase 4 | 11704 | -0.065 | -0.4420 | Yes |
| 65 | KIF5B | KIF5B Entrez, Source | kinesin family member 5B | 11786 | -0.067 | -0.4322 | Yes |
| 66 | NAP1L1 | NAP1L1 Entrez, Source | nucleosome assembly protein 1-like 1 | 11929 | -0.071 | -0.4260 | Yes |
| 67 | BZW2 | BZW2 Entrez, Source | basic leucine zipper and W2 domains 2 | 12150 | -0.078 | -0.4240 | Yes |
| 68 | DEK | DEK Entrez, Source | DEK oncogene (DNA binding) | 12158 | -0.078 | -0.4059 | Yes |
| 69 | ANXA4 | ANXA4 Entrez, Source | annexin A4 | 12281 | -0.082 | -0.3956 | Yes |
| 70 | DUSP18 | DUSP18 Entrez, Source | dual specificity phosphatase 18 | 12828 | -0.113 | -0.4098 | Yes |
| 71 | TMSB4X | TMSB4X Entrez, Source | thymosin, beta 4, X-linked | 12959 | -0.127 | -0.3893 | Yes |
| 72 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 13194 | -0.185 | -0.3627 | Yes |
| 73 | MCM6 | MCM6 Entrez, Source | MCM6 minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) | 13196 | -0.186 | -0.3184 | Yes |
| 74 | KIAA0101 | KIAA0101 Entrez, Source | KIAA0101 | 13232 | -0.213 | -0.2701 | Yes |
| 75 | CLIC1 | CLIC1 Entrez, Source | chloride intracellular channel 1 | 13242 | -0.218 | -0.2188 | Yes |
| 76 | ANXA3 | ANXA3 Entrez, Source | annexin A3 | 13255 | -0.224 | -0.1661 | Yes |
| 77 | SERPINH1 | SERPINH1 Entrez, Source | serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) | 13285 | -0.265 | -0.1052 | Yes |
| 78 | S100A6 | S100A6 Entrez, Source | S100 calcium binding protein A6 | 13330 | -0.458 | 0.0009 | Yes |