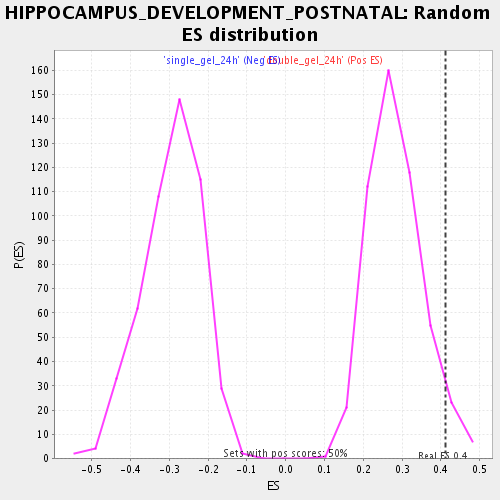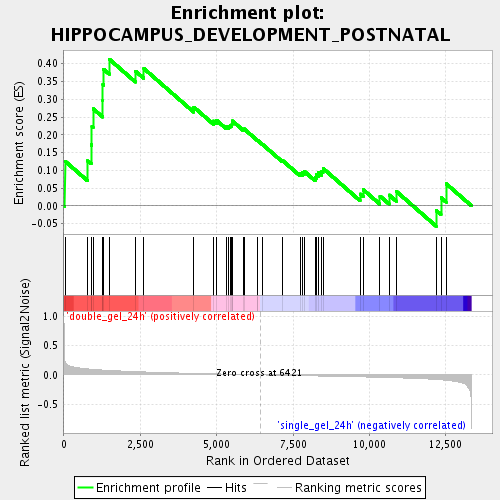
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | HIPPOCAMPUS_DEVELOPMENT_POSTNATAL |
| Enrichment Score (ES) | 0.41284713 |
| Normalized Enrichment Score (NES) | 1.4540539 |
| Nominal p-value | 0.04225352 |
| FDR q-value | 0.29766545 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EGR1 | EGR1 Entrez, Source | early growth response 1 | 35 | 0.229 | 0.1261 | Yes |
| 2 | KCNMA1 | KCNMA1 Entrez, Source | potassium large conductance calcium-activated channel, subfamily M, alpha member 1 | 777 | 0.100 | 0.1270 | Yes |
| 3 | CX3CL1 | CX3CL1 Entrez, Source | chemokine (C-X3-C motif) ligand 1 | 894 | 0.096 | 0.1721 | Yes |
| 4 | RYR2 | RYR2 Entrez, Source | ryanodine receptor 2 (cardiac) | 917 | 0.095 | 0.2238 | Yes |
| 5 | GRIA2 | GRIA2 Entrez, Source | glutamate receptor, ionotropic, AMPA 2 | 957 | 0.093 | 0.2734 | Yes |
| 6 | VAMP2 | VAMP2 Entrez, Source | vesicle-associated membrane protein 2 (synaptobrevin 2) | 1269 | 0.081 | 0.2958 | Yes |
| 7 | EEF1A2 | EEF1A2 Entrez, Source | eukaryotic translation elongation factor 1 alpha 2 | 1275 | 0.081 | 0.3411 | Yes |
| 8 | PYGM | PYGM Entrez, Source | phosphorylase, glycogen; muscle (McArdle syndrome, glycogen storage disease type V) | 1297 | 0.080 | 0.3847 | Yes |
| 9 | SOX10 | SOX10 Entrez, Source | SRY (sex determining region Y)-box 10 | 1485 | 0.075 | 0.4128 | Yes |
| 10 | GEFT | GEFT Entrez, Source | - | 2342 | 0.055 | 0.3792 | No |
| 11 | NRGN | NRGN Entrez, Source | neurogranin (protein kinase C substrate, RC3) | 2616 | 0.049 | 0.3866 | No |
| 12 | PFKL | PFKL Entrez, Source | phosphofructokinase, liver | 4248 | 0.025 | 0.2782 | No |
| 13 | ATP6V0C | ATP6V0C Entrez, Source | ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c | 4900 | 0.017 | 0.2390 | No |
| 14 | GRIA1 | GRIA1 Entrez, Source | glutamate receptor, ionotropic, AMPA 1 | 4994 | 0.016 | 0.2410 | No |
| 15 | MDH1 | MDH1 Entrez, Source | malate dehydrogenase 1, NAD (soluble) | 5310 | 0.012 | 0.2243 | No |
| 16 | NAPA | NAPA Entrez, Source | N-ethylmaleimide-sensitive factor attachment protein, alpha | 5398 | 0.011 | 0.2242 | No |
| 17 | STXBP1 | STXBP1 Entrez, Source | syntaxin binding protein 1 | 5437 | 0.011 | 0.2274 | No |
| 18 | ALDOA | ALDOA Entrez, Source | aldolase A, fructose-bisphosphate | 5499 | 0.010 | 0.2286 | No |
| 19 | COX8A | COX8A Entrez, Source | cytochrome c oxidase subunit 8A (ubiquitous) | 5505 | 0.010 | 0.2339 | No |
| 20 | IGHMBP2 | IGHMBP2 Entrez, Source | immunoglobulin mu binding protein 2 | 5510 | 0.010 | 0.2393 | No |
| 21 | SH3GL2 | SH3GL2 Entrez, Source | SH3-domain GRB2-like 2 | 5874 | 0.006 | 0.2153 | No |
| 22 | RAB3A | RAB3A Entrez, Source | RAB3A, member RAS oncogene family | 5896 | 0.006 | 0.2168 | No |
| 23 | TPI1 | TPI1 Entrez, Source | triosephosphate isomerase 1 | 6341 | 0.001 | 0.1839 | No |
| 24 | ATP6V1C1 | ATP6V1C1 Entrez, Source | ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1 | 6343 | 0.001 | 0.1843 | No |
| 25 | GPI | GPI Entrez, Source | glucose phosphate isomerase | 6497 | -0.001 | 0.1732 | No |
| 26 | ATP2A2 | ATP2A2 Entrez, Source | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | 7144 | -0.007 | 0.1286 | No |
| 27 | BIN1 | BIN1 Entrez, Source | bridging integrator 1 | 7729 | -0.013 | 0.0919 | No |
| 28 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 7807 | -0.014 | 0.0938 | No |
| 29 | BDNF | BDNF Entrez, Source | brain-derived neurotrophic factor | 7868 | -0.014 | 0.0973 | No |
| 30 | GRINA | GRINA Entrez, Source | glutamate receptor, ionotropic, N-methyl D-asparate-associated protein 1 (glutamate binding) | 8227 | -0.018 | 0.0806 | No |
| 31 | PRDX6 | PRDX6 Entrez, Source | peroxiredoxin 6 | 8269 | -0.018 | 0.0878 | No |
| 32 | AIP | AIP Entrez, Source | aryl hydrocarbon receptor interacting protein | 8329 | -0.019 | 0.0941 | No |
| 33 | VDAC1 | VDAC1 Entrez, Source | voltage-dependent anion channel 1 | 8435 | -0.020 | 0.0975 | No |
| 34 | DGKZ | DGKZ Entrez, Source | diacylglycerol kinase, zeta 104kDa | 8481 | -0.020 | 0.1056 | No |
| 35 | CLTB | CLTB Entrez, Source | clathrin, light chain (Lcb) | 9694 | -0.034 | 0.0336 | No |
| 36 | PPP3CA | PPP3CA Entrez, Source | protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | 9796 | -0.035 | 0.0458 | No |
| 37 | ENO2 | ENO2 Entrez, Source | enolase 2 (gamma, neuronal) | 10342 | -0.041 | 0.0279 | No |
| 38 | NDUFS2 | NDUFS2 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase) | 10644 | -0.045 | 0.0308 | No |
| 39 | NTSR2 | NTSR2 Entrez, Source | neurotensin receptor 2 | 10880 | -0.049 | 0.0409 | No |
| 40 | MAPK3 | MAPK3 Entrez, Source | mitogen-activated protein kinase 3 | 12184 | -0.079 | -0.0127 | No |
| 41 | TYRO3 | TYRO3 Entrez, Source | TYRO3 protein tyrosine kinase | 12342 | -0.085 | 0.0233 | No |
| 42 | LDHB | LDHB Entrez, Source | lactate dehydrogenase B | 12511 | -0.092 | 0.0625 | No |