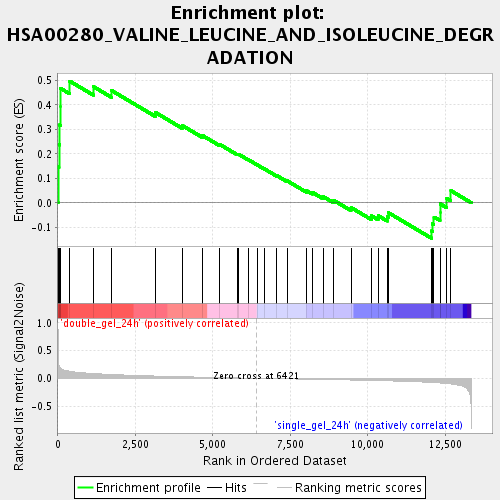
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | HSA00280_VALINE_LEUCINE_AND_ISOLEUCINE_DEGRADATION |
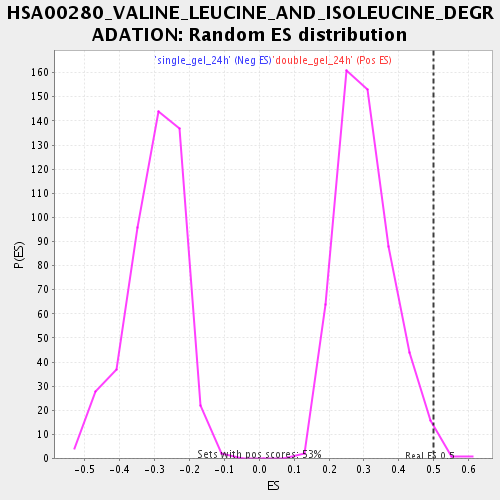
| Enrichment Score (ES) | 0.49759865 |
| Normalized Enrichment Score (NES) | 1.6499841 |
| Nominal p-value | 0.00754717 |
| FDR q-value | 0.16134332 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 7 | 0.362 | 0.1467 | Yes |
| 2 | HMGCS1 | HMGCS1 Entrez, Source | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 (soluble) | 36 | 0.229 | 0.2376 | Yes |
| 3 | ACAT2 | ACAT2 Entrez, Source | acetyl-Coenzyme A acetyltransferase 2 (acetoacetyl Coenzyme A thiolase) | 57 | 0.202 | 0.3183 | Yes |
| 4 | HMGCS2 | HMGCS2 Entrez, Source | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 (mitochondrial) | 80 | 0.187 | 0.3926 | Yes |
| 5 | ACAA2 | ACAA2 Entrez, Source | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | 83 | 0.185 | 0.4679 | Yes |
| 6 | HADH | HADH Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase | 379 | 0.127 | 0.4976 | Yes |
| 7 | BCAT1 | BCAT1 Entrez, Source | branched chain aminotransferase 1, cytosolic | 1144 | 0.086 | 0.4751 | No |
| 8 | ACAT1 | ACAT1 Entrez, Source | acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | 1732 | 0.069 | 0.4590 | No |
| 9 | AOX1 | AOX1 Entrez, Source | aldehyde oxidase 1 | 3148 | 0.041 | 0.3693 | No |
| 10 | BCAT2 | BCAT2 Entrez, Source | branched chain aminotransferase 2, mitochondrial | 4011 | 0.028 | 0.3160 | No |
| 11 | ACADM | ACADM Entrez, Source | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain | 4664 | 0.020 | 0.2752 | No |
| 12 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 5228 | 0.013 | 0.2383 | No |
| 13 | HADHA | HADHA Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 5790 | 0.007 | 0.1989 | No |
| 14 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 5819 | 0.007 | 0.1995 | No |
| 15 | ACADS | ACADS Entrez, Source | acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain | 6143 | 0.003 | 0.1765 | No |
| 16 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 6435 | -0.000 | 0.1547 | No |
| 17 | HSD17B4 | HSD17B4 Entrez, Source | hydroxysteroid (17-beta) dehydrogenase 4 | 6653 | -0.002 | 0.1394 | No |
| 18 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 7066 | -0.006 | 0.1110 | No |
| 19 | HMGCL | HMGCL Entrez, Source | 3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase (hydroxymethylglutaricaciduria) | 7395 | -0.009 | 0.0902 | No |
| 20 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 8031 | -0.016 | 0.0490 | No |
| 21 | ALDH1B1 | ALDH1B1 Entrez, Source | aldehyde dehydrogenase 1 family, member B1 | 8208 | -0.018 | 0.0430 | No |
| 22 | PCCB | PCCB Entrez, Source | propionyl Coenzyme A carboxylase, beta polypeptide | 8561 | -0.021 | 0.0252 | No |
| 23 | ABAT | ABAT Entrez, Source | 4-aminobutyrate aminotransferase | 8902 | -0.025 | 0.0098 | No |
| 24 | DLD | DLD Entrez, Source | dihydrolipoamide dehydrogenase | 9461 | -0.031 | -0.0195 | No |
| 25 | HIBADH | HIBADH Entrez, Source | 3-hydroxyisobutyrate dehydrogenase | 10119 | -0.039 | -0.0532 | No |
| 26 | DBT | DBT Entrez, Source | dihydrolipoamide branched chain transacylase E2 | 10335 | -0.041 | -0.0527 | No |
| 27 | ALDH6A1 | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 10628 | -0.045 | -0.0562 | No |
| 28 | HIBCH | HIBCH Entrez, Source | 3-hydroxyisobutyryl-Coenzyme A hydrolase | 10662 | -0.046 | -0.0401 | No |
| 29 | BCKDHA | BCKDHA Entrez, Source | branched chain keto acid dehydrogenase E1, alpha polypeptide | 12065 | -0.075 | -0.1151 | No |
| 30 | ALDH7A1 | ALDH7A1 Entrez, Source | aldehyde dehydrogenase 7 family, member A1 | 12088 | -0.076 | -0.0860 | No |
| 31 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 12137 | -0.077 | -0.0582 | No |
| 32 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 12333 | -0.084 | -0.0385 | No |
| 33 | MCCC1 | MCCC1 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 1 (alpha) | 12336 | -0.085 | -0.0042 | No |
| 34 | MCCC2 | MCCC2 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 2 (beta) | 12548 | -0.094 | 0.0182 | No |
| 35 | IVD | IVD Entrez, Source | isovaleryl Coenzyme A dehydrogenase | 12683 | -0.102 | 0.0495 | No |