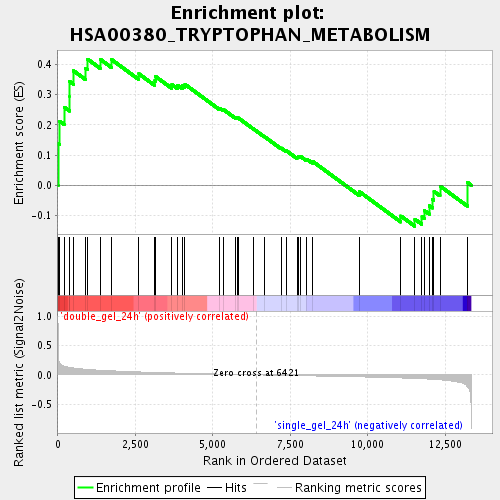
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | HSA00380_TRYPTOPHAN_METABOLISM |
| Enrichment Score (ES) | 0.41722143 |
| Normalized Enrichment Score (NES) | 1.4556957 |
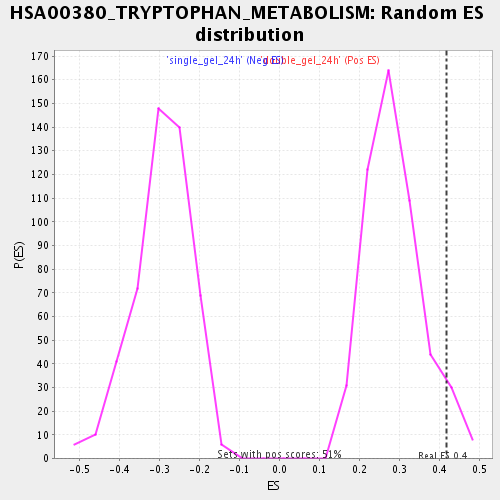
| Nominal p-value | 0.049212597 |
| FDR q-value | 0.2961559 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 7 | 0.362 | 0.1383 | Yes |
| 2 | ACAT2 | ACAT2 Entrez, Source | acetyl-Coenzyme A acetyltransferase 2 (acetoacetyl Coenzyme A thiolase) | 57 | 0.202 | 0.2121 | Yes |
| 3 | CYP1A1 | CYP1A1 Entrez, Source | cytochrome P450, family 1, subfamily A, polypeptide 1 | 204 | 0.149 | 0.2581 | Yes |
| 4 | AOC3 | AOC3 Entrez, Source | amine oxidase, copper containing 3 (vascular adhesion protein 1) | 374 | 0.128 | 0.2946 | Yes |
| 5 | HADH | HADH Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase | 379 | 0.127 | 0.3431 | Yes |
| 6 | TDO2 | TDO2 Entrez, Source | tryptophan 2,3-dioxygenase | 504 | 0.117 | 0.3788 | Yes |
| 7 | CYP1B1 | CYP1B1 Entrez, Source | cytochrome P450, family 1, subfamily B, polypeptide 1 | 895 | 0.096 | 0.3862 | Yes |
| 8 | ABP1 | ABP1 Entrez, Source | amiloride binding protein 1 (amine oxidase (copper-containing)) | 959 | 0.093 | 0.4172 | Yes |
| 9 | AANAT | AANAT Entrez, Source | arylalkylamine N-acetyltransferase | 1366 | 0.078 | 0.4168 | No |
| 10 | ACAT1 | ACAT1 Entrez, Source | acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | 1732 | 0.069 | 0.4158 | No |
| 11 | ASMT | ASMT Entrez, Source | acetylserotonin O-methyltransferase | 2613 | 0.049 | 0.3686 | No |
| 12 | CAT | CAT Entrez, Source | catalase | 3129 | 0.041 | 0.3457 | No |
| 13 | AOX1 | AOX1 Entrez, Source | aldehyde oxidase 1 | 3148 | 0.041 | 0.3601 | No |
| 14 | NFX1 | NFX1 Entrez, Source | nuclear transcription factor, X-box binding 1 | 3678 | 0.033 | 0.3328 | No |
| 15 | DDC | DDC Entrez, Source | dopa decarboxylase (aromatic L-amino acid decarboxylase) | 3861 | 0.030 | 0.3307 | No |
| 16 | HAAO | HAAO Entrez, Source | 3-hydroxyanthranilate 3,4-dioxygenase | 4023 | 0.028 | 0.3294 | No |
| 17 | LCMT1 | LCMT1 Entrez, Source | leucine carboxyl methyltransferase 1 | 4098 | 0.027 | 0.3342 | No |
| 18 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 5228 | 0.013 | 0.2545 | No |
| 19 | INDO | INDO Entrez, Source | indoleamine-pyrrole 2,3 dioxygenase | 5344 | 0.012 | 0.2504 | No |
| 20 | CYP1A2 | CYP1A2 Entrez, Source | cytochrome P450, family 1, subfamily A, polypeptide 2 | 5738 | 0.007 | 0.2237 | No |
| 21 | HADHA | HADHA Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 5790 | 0.007 | 0.2225 | No |
| 22 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 5819 | 0.007 | 0.2229 | No |
| 23 | METTL6 | METTL6 Entrez, Source | methyltransferase like 6 | 6307 | 0.001 | 0.1868 | No |
| 24 | HSD17B4 | HSD17B4 Entrez, Source | hydroxysteroid (17-beta) dehydrogenase 4 | 6653 | -0.002 | 0.1618 | No |
| 25 | CARM1 | CARM1 Entrez, Source | coactivator-associated arginine methyltransferase 1 | 7204 | -0.008 | 0.1234 | No |
| 26 | KMO | KMO Entrez, Source | kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) | 7374 | -0.009 | 0.1142 | No |
| 27 | TPH1 | TPH1 Entrez, Source | tryptophan hydroxylase 1 (tryptophan 5-monooxygenase) | 7719 | -0.013 | 0.0932 | No |
| 28 | AADAT | AADAT Entrez, Source | aminoadipate aminotransferase | 7758 | -0.013 | 0.0954 | No |
| 29 | WARS | WARS Entrez, Source | tryptophanyl-tRNA synthetase | 7827 | -0.014 | 0.0956 | No |
| 30 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 8031 | -0.016 | 0.0865 | No |
| 31 | ALDH1B1 | ALDH1B1 Entrez, Source | aldehyde dehydrogenase 1 family, member B1 | 8208 | -0.018 | 0.0801 | No |
| 32 | PRMT3 | PRMT3 Entrez, Source | protein arginine methyltransferase 3 | 9734 | -0.034 | -0.0214 | No |
| 33 | PRMT2 | PRMT2 Entrez, Source | protein arginine methyltransferase 2 | 11068 | -0.052 | -0.1016 | No |
| 34 | MAOB | MAOB Entrez, Source | monoamine oxidase B | 11520 | -0.061 | -0.1122 | No |
| 35 | MAOA | MAOA Entrez, Source | monoamine oxidase A | 11749 | -0.066 | -0.1041 | No |
| 36 | LCMT2 | LCMT2 Entrez, Source | leucine carboxyl methyltransferase 2 | 11824 | -0.068 | -0.0835 | No |
| 37 | KYNU | KYNU Entrez, Source | kynureninase (L-kynurenine hydrolase) | 11987 | -0.072 | -0.0679 | No |
| 38 | ALDH7A1 | ALDH7A1 Entrez, Source | aldehyde dehydrogenase 7 family, member A1 | 12088 | -0.076 | -0.0464 | No |
| 39 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 12137 | -0.077 | -0.0204 | No |
| 40 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 12333 | -0.084 | -0.0027 | No |
| 41 | ACMSD | ACMSD Entrez, Source | aminocarboxymuconate semialdehyde decarboxylase | 13218 | -0.205 | 0.0093 | No |