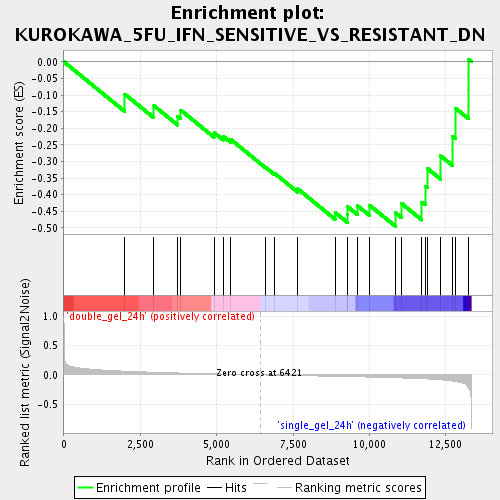
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | single_gel_24h |
| GeneSet | KUROKAWA_5FU_IFN_SENSITIVE_VS_RESISTANT_DN |
| Enrichment Score (ES) | -0.49503788 |
| Normalized Enrichment Score (NES) | -1.571191 |
| Nominal p-value | 0.026584867 |
| FDR q-value | 0.25658163 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IFNB1 | IFNB1 Entrez, Source | interferon, beta 1, fibroblast | 1990 | 0.063 | -0.0971 | No |
| 2 | HK2 | HK2 Entrez, Source | hexokinase 2 | 2945 | 0.044 | -0.1321 | No |
| 3 | EGR3 | EGR3 Entrez, Source | early growth response 3 | 3724 | 0.032 | -0.1637 | No |
| 4 | NR1D2 | NR1D2 Entrez, Source | nuclear receptor subfamily 1, group D, member 2 | 3822 | 0.031 | -0.1454 | No |
| 5 | ERO1L | ERO1L Entrez, Source | ERO1-like (S. cerevisiae) | 4913 | 0.017 | -0.2131 | No |
| 6 | BRCA2 | BRCA2 Entrez, Source | breast cancer 2, early onset | 5213 | 0.014 | -0.2243 | No |
| 7 | PPP2R2A | PPP2R2A Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), alpha isoform | 5448 | 0.011 | -0.2329 | No |
| 8 | EEF2 | EEF2 Entrez, Source | eukaryotic translation elongation factor 2 | 6605 | -0.002 | -0.3180 | No |
| 9 | LENG8 | LENG8 Entrez, Source | leukocyte receptor cluster (LRC) member 8 | 6900 | -0.005 | -0.3362 | No |
| 10 | SND1 | SND1 Entrez, Source | - | 7653 | -0.012 | -0.3826 | No |
| 11 | BIRC4 | BIRC4 Entrez, Source | baculoviral IAP repeat-containing 4 | 8879 | -0.025 | -0.4540 | No |
| 12 | CALM1 | CALM1 Entrez, Source | calmodulin 1 (phosphorylase kinase, delta) | 9285 | -0.029 | -0.4603 | Yes |
| 13 | ATP6V1D | ATP6V1D Entrez, Source | ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D | 9292 | -0.029 | -0.4367 | Yes |
| 14 | CLNS1A | CLNS1A Entrez, Source | chloride channel, nucleotide-sensitive, 1A | 9598 | -0.033 | -0.4324 | Yes |
| 15 | CTPS2 | CTPS2 Entrez, Source | CTP synthase II | 10001 | -0.037 | -0.4317 | Yes |
| 16 | BLMH | BLMH Entrez, Source | bleomycin hydrolase | 10846 | -0.049 | -0.4544 | Yes |
| 17 | UNC5B | UNC5B Entrez, Source | unc-5 homolog B (C. elegans) | 11046 | -0.052 | -0.4263 | Yes |
| 18 | S100A10 | S100A10 Entrez, Source | S100 calcium binding protein A10 | 11713 | -0.065 | -0.4221 | Yes |
| 19 | GLS | GLS Entrez, Source | glutaminase | 11830 | -0.068 | -0.3741 | Yes |
| 20 | ENPEP | ENPEP Entrez, Source | glutamyl aminopeptidase (aminopeptidase A) | 11892 | -0.070 | -0.3206 | Yes |
| 21 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 12333 | -0.084 | -0.2834 | Yes |
| 22 | PDCD4 | PDCD4 Entrez, Source | programmed cell death 4 (neoplastic transformation inhibitor) | 12722 | -0.105 | -0.2254 | Yes |
| 23 | TMPO | TMPO Entrez, Source | thymopoietin | 12812 | -0.111 | -0.1395 | Yes |
| 24 | PVRL2 | PVRL2 Entrez, Source | poliovirus receptor-related 2 (herpesvirus entry mediator B) | 13237 | -0.215 | 0.0079 | Yes |