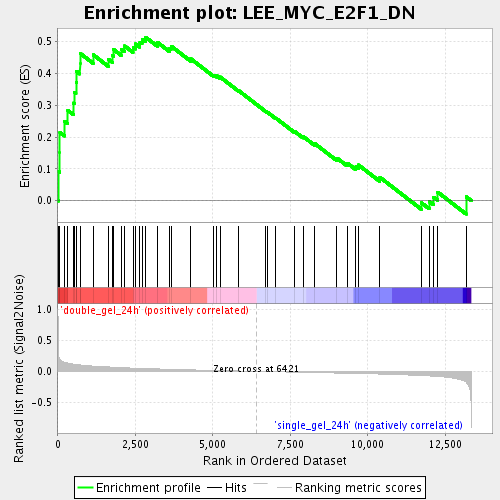
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | LEE_MYC_E2F1_DN |
| Enrichment Score (ES) | 0.5132161 |
| Normalized Enrichment Score (NES) | 1.8253667 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.08142963 |
| FWER p-Value | 0.953 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | G6PC | G6PC Entrez, Source | glucose-6-phosphatase, catalytic subunit | 14 | 0.295 | 0.0923 | Yes |
| 2 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 59 | 0.201 | 0.1525 | Yes |
| 3 | FABP1 | FABP1 Entrez, Source | fatty acid binding protein 1, liver | 63 | 0.197 | 0.2143 | Yes |
| 4 | G0S2 | G0S2 Entrez, Source | G0/G1switch 2 | 215 | 0.147 | 0.2493 | Yes |
| 5 | BHMT | BHMT Entrez, Source | betaine-homocysteine methyltransferase | 317 | 0.133 | 0.2838 | Yes |
| 6 | TDO2 | TDO2 Entrez, Source | tryptophan 2,3-dioxygenase | 504 | 0.117 | 0.3069 | Yes |
| 7 | MAT1A | MAT1A Entrez, Source | methionine adenosyltransferase I, alpha | 530 | 0.115 | 0.3413 | Yes |
| 8 | RGS16 | RGS16 Entrez, Source | regulator of G-protein signalling 16 | 584 | 0.111 | 0.3724 | Yes |
| 9 | CYP2E1 | CYP2E1 Entrez, Source | cytochrome P450, family 2, subfamily E, polypeptide 1 | 601 | 0.110 | 0.4060 | Yes |
| 10 | C4BPA | C4BPA Entrez, Source | complement component 4 binding protein, alpha | 712 | 0.104 | 0.4305 | Yes |
| 11 | ELA2 | ELA2 Entrez, Source | elastase 2, neutrophil | 716 | 0.103 | 0.4629 | Yes |
| 12 | SLC25A22 | SLC25A22 Entrez, Source | solute carrier family 25 (mitochondrial carrier: glutamate), member 22 | 1139 | 0.086 | 0.4583 | Yes |
| 13 | TRPV2 | TRPV2 Entrez, Source | transient receptor potential cation channel, subfamily V, member 2 | 1634 | 0.072 | 0.4438 | Yes |
| 14 | ACSL1 | ACSL1 Entrez, Source | acyl-CoA synthetase long-chain family member 1 | 1761 | 0.068 | 0.4559 | Yes |
| 15 | CYP7B1 | CYP7B1 Entrez, Source | cytochrome P450, family 7, subfamily B, polypeptide 1 | 1796 | 0.068 | 0.4747 | Yes |
| 16 | HSD11B1 | HSD11B1 Entrez, Source | hydroxysteroid (11-beta) dehydrogenase 1 | 2044 | 0.061 | 0.4755 | Yes |
| 17 | OTC | OTC Entrez, Source | ornithine carbamoyltransferase | 2152 | 0.059 | 0.4860 | Yes |
| 18 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 2425 | 0.053 | 0.4824 | Yes |
| 19 | ADIPOR2 | ADIPOR2 Entrez, Source | adiponectin receptor 2 | 2509 | 0.051 | 0.4924 | Yes |
| 20 | SFTPD | SFTPD Entrez, Source | surfactant, pulmonary-associated protein D | 2645 | 0.049 | 0.4977 | Yes |
| 21 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 2716 | 0.048 | 0.5075 | Yes |
| 22 | ELOVL5 | ELOVL5 Entrez, Source | ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast) | 2833 | 0.046 | 0.5132 | Yes |
| 23 | EPHX2 | EPHX2 Entrez, Source | epoxide hydrolase 2, cytoplasmic | 3215 | 0.040 | 0.4971 | No |
| 24 | PXMP2 | PXMP2 Entrez, Source | peroxisomal membrane protein 2, 22kDa | 3604 | 0.034 | 0.4786 | No |
| 25 | HBP1 | HBP1 Entrez, Source | HMG-box transcription factor 1 | 3652 | 0.033 | 0.4855 | No |
| 26 | IGF1 | IGF1 Entrez, Source | insulin-like growth factor 1 (somatomedin C) | 4287 | 0.025 | 0.4457 | No |
| 27 | IGFBP2 | IGFBP2 Entrez, Source | insulin-like growth factor binding protein 2, 36kDa | 5034 | 0.016 | 0.3945 | No |
| 28 | HAL | HAL Entrez, Source | histidine ammonia-lyase | 5120 | 0.015 | 0.3928 | No |
| 29 | CYP4F2 | CYP4F2 Entrez, Source | cytochrome P450, family 4, subfamily F, polypeptide 2 | 5234 | 0.013 | 0.3885 | No |
| 30 | ALAD | ALAD Entrez, Source | aminolevulinate, delta-, dehydratase | 5837 | 0.006 | 0.3452 | No |
| 31 | CTRC | CTRC Entrez, Source | chymotrypsin C (caldecrin) | 6708 | -0.003 | 0.2807 | No |
| 32 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 6761 | -0.003 | 0.2779 | No |
| 33 | CPOX | CPOX Entrez, Source | coproporphyrinogen oxidase | 7027 | -0.006 | 0.2598 | No |
| 34 | NR1H3 | NR1H3 Entrez, Source | nuclear receptor subfamily 1, group H, member 3 | 7622 | -0.012 | 0.2189 | No |
| 35 | OAT | OAT Entrez, Source | ornithine aminotransferase (gyrate atrophy) | 7916 | -0.015 | 0.2015 | No |
| 36 | ABCD3 | ABCD3 Entrez, Source | ATP-binding cassette, sub-family D (ALD), member 3 | 8286 | -0.019 | 0.1796 | No |
| 37 | PNLIPRP1 | PNLIPRP1 Entrez, Source | pancreatic lipase-related protein 1 | 9006 | -0.026 | 0.1338 | No |
| 38 | MAPKAPK2 | MAPKAPK2 Entrez, Source | mitogen-activated protein kinase-activated protein kinase 2 | 9335 | -0.029 | 0.1184 | No |
| 39 | RARRES1 | RARRES1 Entrez, Source | retinoic acid receptor responder (tazarotene induced) 1 | 9619 | -0.033 | 0.1075 | No |
| 40 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 9698 | -0.034 | 0.1124 | No |
| 41 | GHR | GHR Entrez, Source | growth hormone receptor | 10389 | -0.042 | 0.0737 | No |
| 42 | BAAT | BAAT Entrez, Source | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 11730 | -0.065 | -0.0064 | No |
| 43 | KYNU | KYNU Entrez, Source | kynureninase (L-kynurenine hydrolase) | 11987 | -0.072 | -0.0027 | No |
| 44 | IL1RAP | IL1RAP Entrez, Source | interleukin 1 receptor accessory protein | 12123 | -0.077 | 0.0114 | No |
| 45 | FABP2 | FABP2 Entrez, Source | fatty acid binding protein 2, intestinal | 12256 | -0.081 | 0.0271 | No |
| 46 | RGN | RGN Entrez, Source | regucalcin (senescence marker protein-30) | 13176 | -0.172 | 0.0125 | No |