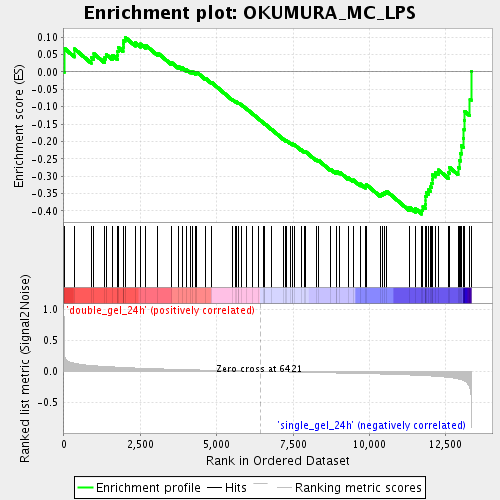
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | single_gel_24h |
| GeneSet | OKUMURA_MC_LPS |
| Enrichment Score (ES) | -0.4091008 |
| Normalized Enrichment Score (NES) | -1.650546 |
| Nominal p-value | 0.003976143 |
| FDR q-value | 0.19302206 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PHLDA1 | PHLDA1 Entrez, Source | pleckstrin homology-like domain, family A, member 1 | 6 | 0.386 | 0.0688 | No |
| 2 | CCL5 | CCL5 Entrez, Source | chemokine (C-C motif) ligand 5 | 347 | 0.130 | 0.0665 | No |
| 3 | IGFBP5 | IGFBP5 Entrez, Source | insulin-like growth factor binding protein 5 | 892 | 0.096 | 0.0427 | No |
| 4 | MSX2 | MSX2 Entrez, Source | msh homeobox homolog 2 (Drosophila) | 981 | 0.092 | 0.0526 | No |
| 5 | CENTA1 | CENTA1 Entrez, Source | centaurin, alpha 1 | 1328 | 0.079 | 0.0407 | No |
| 6 | VDR | VDR Entrez, Source | vitamin D (1,25- dihydroxyvitamin D3) receptor | 1377 | 0.078 | 0.0511 | No |
| 7 | SPHK1 | SPHK1 Entrez, Source | sphingosine kinase 1 | 1584 | 0.073 | 0.0486 | No |
| 8 | TNFRSF8 | TNFRSF8 Entrez, Source | tumor necrosis factor receptor superfamily, member 8 | 1748 | 0.069 | 0.0487 | No |
| 9 | ACSL1 | ACSL1 Entrez, Source | acyl-CoA synthetase long-chain family member 1 | 1761 | 0.068 | 0.0601 | No |
| 10 | RUNX2 | RUNX2 Entrez, Source | runt-related transcription factor 2 | 1784 | 0.068 | 0.0706 | No |
| 11 | BCL2L11 | BCL2L11 Entrez, Source | BCL2-like 11 (apoptosis facilitator) | 1935 | 0.064 | 0.0707 | No |
| 12 | RGS9 | RGS9 Entrez, Source | regulator of G-protein signalling 9 | 1947 | 0.064 | 0.0813 | No |
| 13 | TNFSF4 | TNFSF4 Entrez, Source | tumor necrosis factor (ligand) superfamily, member 4 (tax-transcriptionally activated glycoprotein 1, 34kDa) | 1960 | 0.063 | 0.0918 | No |
| 14 | BAALC | BAALC Entrez, Source | brain and acute leukemia, cytoplasmic | 2001 | 0.063 | 0.1000 | No |
| 15 | SDC4 | SDC4 Entrez, Source | syndecan 4 (amphiglycan, ryudocan) | 2339 | 0.055 | 0.0844 | No |
| 16 | ADIPOR2 | ADIPOR2 Entrez, Source | adiponectin receptor 2 | 2509 | 0.051 | 0.0809 | No |
| 17 | RASGRP1 | RASGRP1 Entrez, Source | RAS guanyl releasing protein 1 (calcium and DAG-regulated) | 2676 | 0.048 | 0.0771 | No |
| 18 | CD200 | CD200 Entrez, Source | CD200 molecule | 3078 | 0.042 | 0.0543 | No |
| 19 | NUPL1 | NUPL1 Entrez, Source | nucleoporin like 1 | 3508 | 0.035 | 0.0282 | No |
| 20 | GCKR | GCKR Entrez, Source | glucokinase (hexokinase 4) regulator | 3757 | 0.032 | 0.0152 | No |
| 21 | PPIF | PPIF Entrez, Source | peptidylprolyl isomerase F (cyclophilin F) | 3868 | 0.030 | 0.0123 | No |
| 22 | ATF5 | ATF5 Entrez, Source | activating transcription factor 5 | 4006 | 0.028 | 0.0071 | No |
| 23 | PELP1 | PELP1 Entrez, Source | proline, glutamic acid and leucine rich protein 1 | 4140 | 0.026 | 0.0018 | No |
| 24 | CEBPD | CEBPD Entrez, Source | CCAAT/enhancer binding protein (C/EBP), delta | 4222 | 0.026 | 0.0002 | No |
| 25 | ARG2 | ARG2 Entrez, Source | arginase, type II | 4319 | 0.024 | -0.0026 | No |
| 26 | AGT | AGT Entrez, Source | angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | 4353 | 0.024 | -0.0008 | No |
| 27 | AMACR | AMACR Entrez, Source | alpha-methylacyl-CoA racemase | 4628 | 0.020 | -0.0179 | No |
| 28 | GULP1 | GULP1 Entrez, Source | GULP, engulfment adaptor PTB domain containing 1 | 4845 | 0.018 | -0.0309 | No |
| 29 | SLC11A2 | SLC11A2 Entrez, Source | solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 | 5523 | 0.010 | -0.0802 | No |
| 30 | CHST1 | CHST1 Entrez, Source | carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | 5606 | 0.009 | -0.0848 | No |
| 31 | PTGER3 | PTGER3 Entrez, Source | prostaglandin E receptor 3 (subtype EP3) | 5634 | 0.009 | -0.0853 | No |
| 32 | SAFB | SAFB Entrez, Source | scaffold attachment factor B | 5711 | 0.008 | -0.0896 | No |
| 33 | RSN | RSN Entrez, Source | restin (Reed-Steinberg cell-expressed intermediate filament-associated protein) | 5729 | 0.008 | -0.0895 | No |
| 34 | UBE2B | UBE2B Entrez, Source | ubiquitin-conjugating enzyme E2B (RAD6 homolog) | 5812 | 0.007 | -0.0945 | No |
| 35 | PDZD2 | PDZD2 Entrez, Source | PDZ domain containing 2 | 5985 | 0.005 | -0.1066 | No |
| 36 | SCN9A | SCN9A Entrez, Source | sodium channel, voltage-gated, type IX, alpha | 6177 | 0.003 | -0.1206 | No |
| 37 | RAB31 | RAB31 Entrez, Source | RAB31, member RAS oncogene family | 6361 | 0.001 | -0.1343 | No |
| 38 | STAT5A | STAT5A Entrez, Source | signal transducer and activator of transcription 5A | 6535 | -0.001 | -0.1471 | No |
| 39 | EEF1G | EEF1G Entrez, Source | eukaryotic translation elongation factor 1 gamma | 6578 | -0.002 | -0.1500 | No |
| 40 | RCHY1 | RCHY1 Entrez, Source | ring finger and CHY zinc finger domain containing 1 | 6580 | -0.002 | -0.1497 | No |
| 41 | RAB9 | RAB9 Entrez, Source | RAB9, member RAS oncogene family | 6803 | -0.004 | -0.1658 | No |
| 42 | THOC1 | THOC1 Entrez, Source | THO complex 1 | 7179 | -0.007 | -0.1928 | No |
| 43 | AKT3 | AKT3 Entrez, Source | v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma) | 7266 | -0.008 | -0.1978 | No |
| 44 | TSN | TSN Entrez, Source | translin | 7284 | -0.008 | -0.1976 | No |
| 45 | HBLD2 | HBLD2 Entrez, Source | HESB like domain containing 2 | 7407 | -0.010 | -0.2050 | No |
| 46 | HAMP | HAMP Entrez, Source | hepcidin antimicrobial peptide | 7478 | -0.010 | -0.2085 | No |
| 47 | ABCE1 | ABCE1 Entrez, Source | ATP-binding cassette, sub-family E (OABP), member 1 | 7480 | -0.010 | -0.2067 | No |
| 48 | PPAP2B | PPAP2B Entrez, Source | phosphatidic acid phosphatase type 2B | 7532 | -0.011 | -0.2086 | No |
| 49 | RPS21 | RPS21 Entrez, Source | ribosomal protein S21 | 7759 | -0.013 | -0.2233 | No |
| 50 | TNIP2 | TNIP2 Entrez, Source | TNFAIP3 interacting protein 2 | 7875 | -0.014 | -0.2294 | No |
| 51 | ACTA2 | ACTA2 Entrez, Source | actin, alpha 2, smooth muscle, aorta | 7897 | -0.014 | -0.2284 | No |
| 52 | GDF11 | GDF11 Entrez, Source | growth differentiation factor 11 | 8265 | -0.018 | -0.2528 | No |
| 53 | FKBP1A | FKBP1A Entrez, Source | FK506 binding protein 1A, 12kDa | 8328 | -0.019 | -0.2540 | No |
| 54 | ORC6L | ORC6L Entrez, Source | origin recognition complex, subunit 6 like (yeast) | 8724 | -0.023 | -0.2797 | No |
| 55 | CCNB2 | CCNB2 Entrez, Source | cyclin B2 | 8905 | -0.025 | -0.2888 | No |
| 56 | PTGER4 | PTGER4 Entrez, Source | prostaglandin E receptor 4 (subtype EP4) | 8928 | -0.025 | -0.2859 | No |
| 57 | IL5 | IL5 Entrez, Source | interleukin 5 (colony-stimulating factor, eosinophil) | 9032 | -0.026 | -0.2890 | No |
| 58 | LETM1 | LETM1 Entrez, Source | leucine zipper-EF-hand containing transmembrane protein 1 | 9304 | -0.029 | -0.3042 | No |
| 59 | MTM1 | MTM1 Entrez, Source | myotubularin 1 | 9464 | -0.031 | -0.3106 | No |
| 60 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 9698 | -0.034 | -0.3221 | No |
| 61 | NCOA3 | NCOA3 Entrez, Source | nuclear receptor coactivator 3 | 9861 | -0.036 | -0.3279 | No |
| 62 | SGPL1 | SGPL1 Entrez, Source | sphingosine-1-phosphate lyase 1 | 9899 | -0.036 | -0.3242 | No |
| 63 | RAB38 | RAB38 Entrez, Source | RAB38, member RAS oncogene family | 10360 | -0.041 | -0.3515 | No |
| 64 | SMC3 | SMC3 Entrez, Source | structural maintenance of chromosomes 3 | 10429 | -0.042 | -0.3491 | No |
| 65 | PEX12 | PEX12 Entrez, Source | peroxisomal biogenesis factor 12 | 10491 | -0.043 | -0.3459 | No |
| 66 | OPTN | OPTN Entrez, Source | optineurin | 10558 | -0.044 | -0.3430 | No |
| 67 | MTF2 | MTF2 Entrez, Source | metal response element binding transcription factor 2 | 11313 | -0.057 | -0.3896 | No |
| 68 | MARCKS | MARCKS Entrez, Source | myristoylated alanine-rich protein kinase C substrate | 11501 | -0.060 | -0.3929 | No |
| 69 | WT1 | WT1 Entrez, Source | Wilms tumor 1 | 11717 | -0.065 | -0.3974 | Yes |
| 70 | MAOA | MAOA Entrez, Source | monoamine oxidase A | 11749 | -0.066 | -0.3879 | Yes |
| 71 | SLC4A7 | SLC4A7 Entrez, Source | solute carrier family 4, sodium bicarbonate cotransporter, member 7 | 11826 | -0.068 | -0.3814 | Yes |
| 72 | CCHCR1 | CCHCR1 Entrez, Source | coiled-coil alpha-helical rod protein 1 | 11837 | -0.068 | -0.3699 | Yes |
| 73 | PAM | PAM Entrez, Source | peptidylglycine alpha-amidating monooxygenase | 11839 | -0.068 | -0.3577 | Yes |
| 74 | SH3BP5 | SH3BP5 Entrez, Source | SH3-domain binding protein 5 (BTK-associated) | 11864 | -0.069 | -0.3472 | Yes |
| 75 | SMAD3 | SMAD3 Entrez, Source | SMAD, mothers against DPP homolog 3 (Drosophila) | 11928 | -0.071 | -0.3392 | Yes |
| 76 | PTPRK | PTPRK Entrez, Source | protein tyrosine phosphatase, receptor type, K | 11989 | -0.073 | -0.3307 | Yes |
| 77 | C1ORF24 | C1ORF24 Entrez, Source | chromosome 1 open reading frame 24 | 12031 | -0.074 | -0.3206 | Yes |
| 78 | FZD6 | FZD6 Entrez, Source | frizzled homolog 6 (Drosophila) | 12054 | -0.074 | -0.3089 | Yes |
| 79 | ACN9 | ACN9 Entrez, Source | ACN9 homolog (S. cerevisiae) | 12063 | -0.075 | -0.2961 | Yes |
| 80 | CYB5A | CYB5A Entrez, Source | cytochrome b5 type A (microsomal) | 12147 | -0.078 | -0.2884 | Yes |
| 81 | TMEM123 | TMEM123 Entrez, Source | transmembrane protein 123 | 12255 | -0.081 | -0.2819 | Yes |
| 82 | SIRPA | SIRPA Entrez, Source | signal-regulatory protein alpha | 12585 | -0.096 | -0.2895 | Yes |
| 83 | TMEM80 | TMEM80 Entrez, Source | transmembrane protein 80 | 12620 | -0.098 | -0.2745 | Yes |
| 84 | CITED2 | CITED2 Entrez, Source | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | 12899 | -0.121 | -0.2738 | Yes |
| 85 | KCTD3 | KCTD3 Entrez, Source | potassium channel tetramerisation domain containing 3 | 12952 | -0.126 | -0.2550 | Yes |
| 86 | FZD3 | FZD3 Entrez, Source | frizzled homolog 3 (Drosophila) | 12973 | -0.128 | -0.2335 | Yes |
| 87 | ALCAM | ALCAM Entrez, Source | activated leukocyte cell adhesion molecule | 13011 | -0.133 | -0.2124 | Yes |
| 88 | BIRC3 | BIRC3 Entrez, Source | baculoviral IAP repeat-containing 3 | 13071 | -0.143 | -0.1911 | Yes |
| 89 | JAK2 | JAK2 Entrez, Source | Janus kinase 2 (a protein tyrosine kinase) | 13089 | -0.147 | -0.1659 | Yes |
| 90 | MARCKSL1 | MARCKSL1 Entrez, Source | MARCKS-like 1 | 13105 | -0.150 | -0.1401 | Yes |
| 91 | RUNX1 | RUNX1 Entrez, Source | runt-related transcription factor 1 (acute myeloid leukemia 1; aml1 oncogene) | 13119 | -0.153 | -0.1136 | Yes |
| 92 | SCD5 | SCD5 Entrez, Source | stearoyl-CoA desaturase 5 | 13289 | -0.269 | -0.0781 | Yes |
| 93 | IL1RL1 | IL1RL1 Entrez, Source | interleukin 1 receptor-like 1 | 13329 | -0.457 | 0.0010 | Yes |