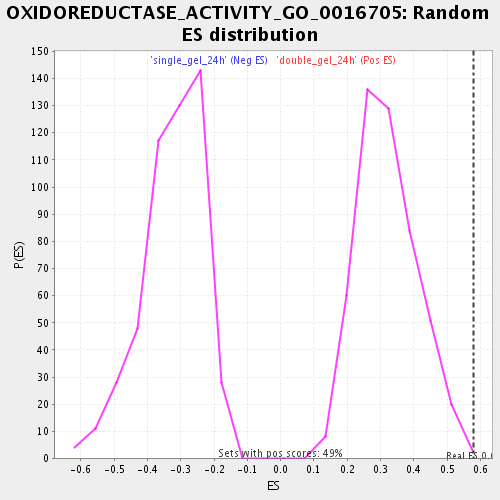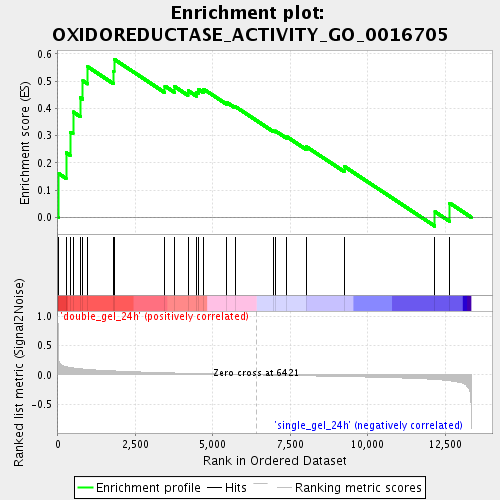
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | OXIDOREDUCTASE_ACTIVITY_GO_0016705 |
| Enrichment Score (ES) | 0.58092636 |
| Normalized Enrichment Score (NES) | 1.8174355 |
| Nominal p-value | 0.0061099795 |
| FDR q-value | 0.08316404 |
| FWER p-Value | 0.966 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CYP26A1 | CYP26A1 Entrez, Source | cytochrome P450, family 26, subfamily A, polypeptide 1 | 27 | 0.239 | 0.1607 | Yes |
| 2 | SC4MOL | SC4MOL Entrez, Source | sterol-C4-methyl oxidase-like | 266 | 0.140 | 0.2378 | Yes |
| 3 | CYP17A1 | CYP17A1 Entrez, Source | cytochrome P450, family 17, subfamily A, polypeptide 1 | 397 | 0.126 | 0.3134 | Yes |
| 4 | NOS3 | NOS3 Entrez, Source | nitric oxide synthase 3 (endothelial cell) | 489 | 0.118 | 0.3871 | Yes |
| 5 | CYP8B1 | CYP8B1 Entrez, Source | cytochrome P450, family 8, subfamily B, polypeptide 1 | 725 | 0.103 | 0.4396 | Yes |
| 6 | HMOX1 | HMOX1 Entrez, Source | heme oxygenase (decycling) 1 | 806 | 0.099 | 0.5012 | Yes |
| 7 | CYP19A1 | CYP19A1 Entrez, Source | cytochrome P450, family 19, subfamily A, polypeptide 1 | 960 | 0.093 | 0.5531 | Yes |
| 8 | CYP7B1 | CYP7B1 Entrez, Source | cytochrome P450, family 7, subfamily B, polypeptide 1 | 1796 | 0.068 | 0.5364 | Yes |
| 9 | CYP11B1 | CYP11B1 Entrez, Source | cytochrome P450, family 11, subfamily B, polypeptide 1 | 1812 | 0.067 | 0.5809 | Yes |
| 10 | P4HA1 | P4HA1 Entrez, Source | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide I | 3453 | 0.036 | 0.4822 | No |
| 11 | PTGS1 | PTGS1 Entrez, Source | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | 3760 | 0.032 | 0.4808 | No |
| 12 | P4HB | P4HB Entrez, Source | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), beta polypeptide | 4199 | 0.026 | 0.4654 | No |
| 13 | SC5DL | SC5DL Entrez, Source | sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, fungal)-like | 4474 | 0.022 | 0.4600 | No |
| 14 | TH | TH Entrez, Source | tyrosine hydroxylase | 4532 | 0.021 | 0.4703 | No |
| 15 | BBOX1 | BBOX1 Entrez, Source | butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 | 4701 | 0.020 | 0.4710 | No |
| 16 | NOS1 | NOS1 Entrez, Source | nitric oxide synthase 1 (neuronal) | 5453 | 0.011 | 0.4219 | No |
| 17 | HMOX2 | HMOX2 Entrez, Source | heme oxygenase (decycling) 2 | 5717 | 0.008 | 0.4074 | No |
| 18 | SCD | SCD Entrez, Source | stearoyl-CoA desaturase (delta-9-desaturase) | 6967 | -0.005 | 0.3172 | No |
| 19 | PLOD3 | PLOD3 Entrez, Source | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 | 7020 | -0.006 | 0.3173 | No |
| 20 | KMO | KMO Entrez, Source | kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) | 7374 | -0.009 | 0.2971 | No |
| 21 | EGLN2 | EGLN2 Entrez, Source | egl nine homolog 2 (C. elegans) | 8020 | -0.016 | 0.2594 | No |
| 22 | EGLN1 | EGLN1 Entrez, Source | egl nine homolog 1 (C. elegans) | 9241 | -0.028 | 0.1871 | No |
| 23 | FADS1 | FADS1 Entrez, Source | fatty acid desaturase 1 | 12169 | -0.078 | 0.0206 | No |
| 24 | PAH | PAH Entrez, Source | phenylalanine hydroxylase | 12643 | -0.099 | 0.0525 | No |