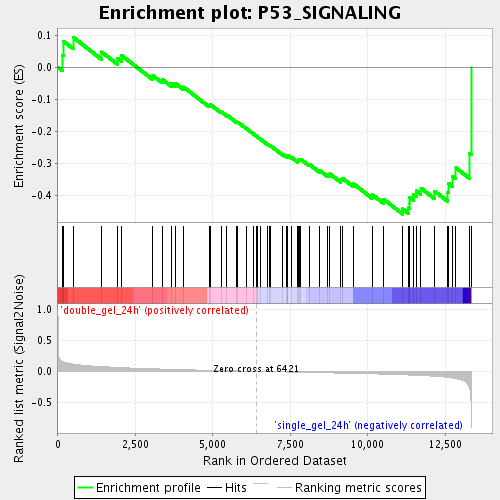
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | single_gel_24h |
| GeneSet | P53_SIGNALING |
| Enrichment Score (ES) | -0.45757928 |
| Normalized Enrichment Score (NES) | -1.6835785 |
| Nominal p-value | 0.001984127 |
| FDR q-value | 0.16810887 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PRKCQ | PRKCQ Entrez, Source | protein kinase C, theta | 147 | 0.160 | 0.0372 | No |
| 2 | CREBBP | CREBBP Entrez, Source | CREB binding protein (Rubinstein-Taybi syndrome) | 179 | 0.152 | 0.0808 | No |
| 3 | CDKN1A | CDKN1A Entrez, Source | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 491 | 0.118 | 0.0931 | No |
| 4 | MYC | MYC Entrez, Source | v-myc myelocytomatosis viral oncogene homolog (avian) | 1411 | 0.077 | 0.0473 | No |
| 5 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 1933 | 0.064 | 0.0274 | No |
| 6 | PRKCB1 | PRKCB1 Entrez, Source | protein kinase C, beta 1 | 2052 | 0.061 | 0.0369 | No |
| 7 | SP1 | SP1 Entrez, Source | Sp1 transcription factor | 3064 | 0.042 | -0.0265 | No |
| 8 | TEAD1 | TEAD1 Entrez, Source | TEA domain family member 1 (SV40 transcriptional enhancer factor) | 3364 | 0.037 | -0.0377 | No |
| 9 | CCNH | CCNH Entrez, Source | cyclin H | 3667 | 0.033 | -0.0505 | No |
| 10 | ESR1 | ESR1 Entrez, Source | estrogen receptor 1 | 3787 | 0.031 | -0.0500 | No |
| 11 | NDRG4 | NDRG4 Entrez, Source | NDRG family member 4 | 4055 | 0.028 | -0.0618 | No |
| 12 | PRKCA | PRKCA Entrez, Source | protein kinase C, alpha | 4883 | 0.018 | -0.1187 | No |
| 13 | THRA | THRA Entrez, Source | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | 4927 | 0.017 | -0.1169 | No |
| 14 | NDRG2 | NDRG2 Entrez, Source | NDRG family member 2 | 5272 | 0.013 | -0.1388 | No |
| 15 | TBP | TBP Entrez, Source | TATA box binding protein | 5455 | 0.011 | -0.1493 | No |
| 16 | BRCA1 | BRCA1 Entrez, Source | breast cancer 1, early onset | 5766 | 0.007 | -0.1705 | No |
| 17 | APEX1 | APEX1 Entrez, Source | APEX nuclease (multifunctional DNA repair enzyme) 1 | 5806 | 0.007 | -0.1714 | No |
| 18 | CSNK2B | CSNK2B Entrez, Source | casein kinase 2, beta polypeptide | 6090 | 0.004 | -0.1916 | No |
| 19 | MAPK8IP2 | MAPK8IP2 Entrez, Source | mitogen-activated protein kinase 8 interacting protein 2 | 6319 | 0.001 | -0.2084 | No |
| 20 | CSNK2A1 | CSNK2A1 Entrez, Source | casein kinase 2, alpha 1 polypeptide | 6396 | 0.000 | -0.2141 | No |
| 21 | NUMB | NUMB Entrez, Source | numb homolog (Drosophila) | 6445 | -0.000 | -0.2176 | No |
| 22 | STAT5A | STAT5A Entrez, Source | signal transducer and activator of transcription 5A | 6535 | -0.001 | -0.2239 | No |
| 23 | FAF1 | FAF1 Entrez, Source | Fas (TNFRSF6) associated factor 1 | 6769 | -0.004 | -0.2404 | No |
| 24 | BCL2 | BCL2 Entrez, Source | B-cell CLL/lymphoma 2 | 6812 | -0.004 | -0.2424 | No |
| 25 | TP53 | TP53 Entrez, Source | tumor protein p53 (Li-Fraumeni syndrome) | 6874 | -0.004 | -0.2457 | No |
| 26 | NFKB1 | NFKB1 Entrez, Source | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (p105) | 7260 | -0.008 | -0.2722 | No |
| 27 | BBC3 | BBC3 Entrez, Source | BCL2 binding component 3 | 7363 | -0.009 | -0.2771 | No |
| 28 | PMP22 | PMP22 Entrez, Source | peripheral myelin protein 22 | 7414 | -0.010 | -0.2779 | No |
| 29 | MAP2K4 | MAP2K4 Entrez, Source | mitogen-activated protein kinase kinase 4 | 7416 | -0.010 | -0.2751 | No |
| 30 | CTSD | CTSD Entrez, Source | cathepsin D (lysosomal aspartyl peptidase) | 7549 | -0.011 | -0.2817 | No |
| 31 | NDRG1 | NDRG1 Entrez, Source | N-myc downstream regulated gene 1 | 7738 | -0.013 | -0.2920 | No |
| 32 | PPIA | PPIA Entrez, Source | peptidylprolyl isomerase A (cyclophilin A) | 7751 | -0.013 | -0.2889 | No |
| 33 | BAX | BAX Entrez, Source | BCL2-associated X protein | 7784 | -0.013 | -0.2873 | No |
| 34 | CSNK1A1 | CSNK1A1 Entrez, Source | casein kinase 1, alpha 1 | 7832 | -0.014 | -0.2866 | No |
| 35 | ACTB | ACTB Entrez, Source | actin, beta | 8105 | -0.017 | -0.3021 | No |
| 36 | TNF | TNF Entrez, Source | tumor necrosis factor (TNF superfamily, member 2) | 8457 | -0.020 | -0.3224 | No |
| 37 | RPL13A | RPL13A Entrez, Source | ribosomal protein L13a | 8692 | -0.023 | -0.3331 | No |
| 38 | CASP9 | CASP9 Entrez, Source | caspase 9, apoptosis-related cysteine peptidase | 8779 | -0.024 | -0.3325 | No |
| 39 | CHEK2 | CHEK2 Entrez, Source | CHK2 checkpoint homolog (S. pombe) | 9129 | -0.027 | -0.3505 | No |
| 40 | RELA | RELA Entrez, Source | v-rel reticuloendotheliosis viral oncogene homolog A, nuclear factor of kappa light polypeptide gene enhancer in B-cells 3, p65 (avian) | 9174 | -0.028 | -0.3454 | No |
| 41 | CDKN2A | CDKN2A Entrez, Source | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 9542 | -0.032 | -0.3634 | No |
| 42 | CDK7 | CDK7 Entrez, Source | cyclin-dependent kinase 7 (MO15 homolog, Xenopus laevis, cdk-activating kinase) | 10156 | -0.039 | -0.3978 | No |
| 43 | EI24 | EI24 Entrez, Source | etoposide induced 2.4 mRNA | 10507 | -0.044 | -0.4110 | No |
| 44 | EP300 | EP300 Entrez, Source | E1A binding protein p300 | 11127 | -0.053 | -0.4415 | Yes |
| 45 | PCAF | PCAF Entrez, Source | p300/CBP-associated factor | 11315 | -0.057 | -0.4382 | Yes |
| 46 | RASA1 | RASA1 Entrez, Source | RAS p21 protein activator (GTPase activating protein) 1 | 11353 | -0.058 | -0.4236 | Yes |
| 47 | E2F1 | E2F1 Entrez, Source | E2F transcription factor 1 | 11355 | -0.058 | -0.4062 | Yes |
| 48 | DAXX | DAXX Entrez, Source | death-associated protein 6 | 11474 | -0.060 | -0.3970 | Yes |
| 49 | HIF1A | HIF1A Entrez, Source | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | 11568 | -0.062 | -0.3853 | Yes |
| 50 | WT1 | WT1 Entrez, Source | Wilms tumor 1 | 11717 | -0.065 | -0.3768 | Yes |
| 51 | FADD | FADD Entrez, Source | Fas (TNFRSF6)-associated via death domain | 12156 | -0.078 | -0.3862 | Yes |
| 52 | SERPINB5 | SERPINB5 Entrez, Source | serpin peptidase inhibitor, clade B (ovalbumin), member 5 | 12574 | -0.095 | -0.3889 | Yes |
| 53 | RB1 | RB1 Entrez, Source | retinoblastoma 1 (including osteosarcoma) | 12607 | -0.097 | -0.3620 | Yes |
| 54 | HSPA4 | HSPA4 Entrez, Source | heat shock 70kDa protein 4 | 12729 | -0.105 | -0.3394 | Yes |
| 55 | CHEK1 | CHEK1 Entrez, Source | CHK1 checkpoint homolog (S. pombe) | 12847 | -0.115 | -0.3136 | Yes |
| 56 | APAF1 | APAF1 Entrez, Source | apoptotic peptidase activating factor | 13280 | -0.257 | -0.2685 | Yes |
| 57 | ACTA1 | ACTA1 Entrez, Source | actin, alpha 1, skeletal muscle | 13342 | -0.905 | -0.0000 | Yes |