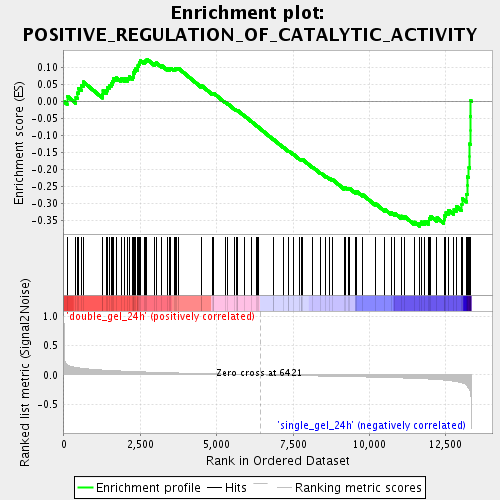
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | single_gel_24h |
| GeneSet | POSITIVE_REGULATION_OF_CATALYTIC_ACTIVITY |
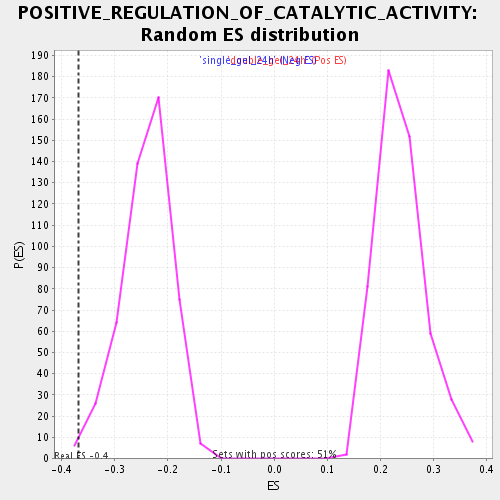
| Enrichment Score (ES) | -0.3676716 |
| Normalized Enrichment Score (NES) | -1.5274898 |
| Nominal p-value | 0.008213553 |
| FDR q-value | 0.2698456 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CHRNA7 | CHRNA7 Entrez, Source | cholinergic receptor, nicotinic, alpha 7 | 107 | 0.175 | 0.0160 | No |
| 2 | EGF | EGF Entrez, Source | epidermal growth factor (beta-urogastrone) | 392 | 0.126 | 0.0119 | No |
| 3 | TNNT2 | TNNT2 Entrez, Source | troponin T type 2 (cardiac) | 441 | 0.122 | 0.0251 | No |
| 4 | CRHR1 | CRHR1 Entrez, Source | corticotropin releasing hormone receptor 1 | 473 | 0.120 | 0.0393 | No |
| 5 | GLP1R | GLP1R Entrez, Source | glucagon-like peptide 1 receptor | 576 | 0.112 | 0.0470 | No |
| 6 | CCKBR | CCKBR Entrez, Source | cholecystokinin B receptor | 632 | 0.108 | 0.0577 | No |
| 7 | CD4 | CD4 Entrez, Source | CD4 molecule | 1266 | 0.081 | 0.0211 | No |
| 8 | CARTPT | CARTPT Entrez, Source | CART prepropeptide | 1278 | 0.081 | 0.0315 | No |
| 9 | ADRA2A | ADRA2A Entrez, Source | adrenergic, alpha-2A-, receptor | 1399 | 0.078 | 0.0331 | No |
| 10 | GADD45B | GADD45B Entrez, Source | growth arrest and DNA-damage-inducible, beta | 1431 | 0.077 | 0.0414 | No |
| 11 | SHC1 | SHC1 Entrez, Source | SHC (Src homology 2 domain containing) transforming protein 1 | 1493 | 0.075 | 0.0471 | No |
| 12 | NF1 | NF1 Entrez, Source | neurofibromin 1 (neurofibromatosis, von Recklinghausen disease, Watson disease) | 1548 | 0.073 | 0.0531 | No |
| 13 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 1596 | 0.073 | 0.0596 | No |
| 14 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 1628 | 0.072 | 0.0671 | No |
| 15 | EDN2 | EDN2 Entrez, Source | endothelin 2 | 1719 | 0.069 | 0.0699 | No |
| 16 | AVPR1B | AVPR1B Entrez, Source | arginine vasopressin receptor 1B | 1870 | 0.066 | 0.0676 | No |
| 17 | IFNB1 | IFNB1 Entrez, Source | interferon, beta 1, fibroblast | 1990 | 0.063 | 0.0673 | No |
| 18 | CHRM1 | CHRM1 Entrez, Source | cholinergic receptor, muscarinic 1 | 2092 | 0.060 | 0.0679 | No |
| 19 | ADRB1 | ADRB1 Entrez, Source | adrenergic, beta-1-, receptor | 2131 | 0.059 | 0.0733 | No |
| 20 | FGF2 | FGF2 Entrez, Source | fibroblast growth factor 2 (basic) | 2253 | 0.056 | 0.0719 | No |
| 21 | GNG3 | GNG3 Entrez, Source | guanine nucleotide binding protein (G protein), gamma 3 | 2275 | 0.056 | 0.0780 | No |
| 22 | BCL2L10 | BCL2L10 Entrez, Source | BCL2-like 10 (apoptosis facilitator) | 2279 | 0.056 | 0.0854 | No |
| 23 | ADRA2C | ADRA2C Entrez, Source | adrenergic, alpha-2C-, receptor | 2313 | 0.055 | 0.0906 | No |
| 24 | TAOK2 | TAOK2 Entrez, Source | TAO kinase 2 | 2341 | 0.055 | 0.0961 | No |
| 25 | GHRL | GHRL Entrez, Source | ghrelin/obestatin preprohormone | 2417 | 0.053 | 0.0977 | No |
| 26 | PARD3 | PARD3 Entrez, Source | par-3 partitioning defective 3 homolog (C. elegans) | 2421 | 0.053 | 0.1049 | No |
| 27 | ERBB2 | ERBB2 Entrez, Source | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | 2456 | 0.053 | 0.1095 | No |
| 28 | C5AR1 | C5AR1 Entrez, Source | complement component 5a receptor 1 | 2477 | 0.052 | 0.1152 | No |
| 29 | CALCR | CALCR Entrez, Source | calcitonin receptor | 2496 | 0.052 | 0.1210 | No |
| 30 | ARHGAP27 | ARHGAP27 Entrez, Source | Rho GTPase activating protein 27 | 2637 | 0.049 | 0.1172 | No |
| 31 | CALCA | CALCA Entrez, Source | calcitonin/calcitonin-related polypeptide, alpha | 2681 | 0.048 | 0.1206 | No |
| 32 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 2716 | 0.048 | 0.1246 | No |
| 33 | ATP7A | ATP7A Entrez, Source | ATPase, Cu++ transporting, alpha polypeptide (Menkes syndrome) | 2970 | 0.043 | 0.1114 | No |
| 34 | GRM4 | GRM4 Entrez, Source | glutamate receptor, metabotropic 4 | 3014 | 0.043 | 0.1141 | No |
| 35 | CAP2 | CAP2 Entrez, Source | CAP, adenylate cyclase-associated protein, 2 (yeast) | 3198 | 0.040 | 0.1058 | No |
| 36 | EREG | EREG Entrez, Source | epiregulin | 3391 | 0.037 | 0.0963 | No |
| 37 | CD24 | CD24 Entrez, Source | CD24 molecule | 3463 | 0.036 | 0.0959 | No |
| 38 | F2 | F2 Entrez, Source | coagulation factor II (thrombin) | 3496 | 0.035 | 0.0984 | No |
| 39 | PRLR | PRLR Entrez, Source | prolactin receptor | 3608 | 0.034 | 0.0946 | No |
| 40 | MADD | MADD Entrez, Source | MAP-kinase activating death domain | 3637 | 0.033 | 0.0971 | No |
| 41 | ADORA3 | ADORA3 Entrez, Source | adenosine A3 receptor | 3680 | 0.033 | 0.0984 | No |
| 42 | ADRB3 | ADRB3 Entrez, Source | adrenergic, beta-3-, receptor | 3743 | 0.032 | 0.0981 | No |
| 43 | AVPR2 | AVPR2 Entrez, Source | arginine vasopressin receptor 2 (nephrogenic diabetes insipidus) | 4489 | 0.022 | 0.0449 | No |
| 44 | LYK5 | LYK5 Entrez, Source | - | 4491 | 0.022 | 0.0478 | No |
| 45 | UBE2N | UBE2N Entrez, Source | ubiquitin-conjugating enzyme E2N (UBC13 homolog, yeast) | 4864 | 0.018 | 0.0222 | No |
| 46 | TGFA | TGFA Entrez, Source | transforming growth factor, alpha | 4880 | 0.018 | 0.0235 | No |
| 47 | DRD5 | DRD5 Entrez, Source | dopamine receptor D5 | 4895 | 0.017 | 0.0248 | No |
| 48 | SERINC1 | SERINC1 Entrez, Source | serine incorporator 1 | 5271 | 0.013 | -0.0017 | No |
| 49 | NMUR2 | NMUR2 Entrez, Source | neuromedin U receptor 2 | 5367 | 0.012 | -0.0073 | No |
| 50 | CENTD2 | CENTD2 Entrez, Source | centaurin, delta 2 | 5588 | 0.009 | -0.0227 | No |
| 51 | EDNRA | EDNRA Entrez, Source | endothelin receptor type A | 5660 | 0.008 | -0.0269 | No |
| 52 | GIPR | GIPR Entrez, Source | gastric inhibitory polypeptide receptor | 5678 | 0.008 | -0.0270 | No |
| 53 | ACR | ACR Entrez, Source | acrosin | 5683 | 0.008 | -0.0262 | No |
| 54 | THY1 | THY1 Entrez, Source | Thy-1 cell surface antigen | 5914 | 0.005 | -0.0429 | No |
| 55 | CCND3 | CCND3 Entrez, Source | cyclin D3 | 6145 | 0.003 | -0.0598 | No |
| 56 | NMUR1 | NMUR1 Entrez, Source | neuromedin U receptor 1 | 6304 | 0.001 | -0.0716 | No |
| 57 | GHRHR | GHRHR Entrez, Source | growth hormone releasing hormone receptor | 6330 | 0.001 | -0.0733 | No |
| 58 | VCP | VCP Entrez, Source | valosin-containing protein | 6382 | 0.000 | -0.0771 | No |
| 59 | PSENEN | PSENEN Entrez, Source | presenilin enhancer 2 homolog (C. elegans) | 6855 | -0.004 | -0.1122 | No |
| 60 | TP53 | TP53 Entrez, Source | tumor protein p53 (Li-Fraumeni syndrome) | 6874 | -0.004 | -0.1130 | No |
| 61 | CXCR4 | CXCR4 Entrez, Source | chemokine (C-X-C motif) receptor 4 | 7175 | -0.007 | -0.1346 | No |
| 62 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7350 | -0.009 | -0.1465 | No |
| 63 | BBC3 | BBC3 Entrez, Source | BCL2 binding component 3 | 7363 | -0.009 | -0.1462 | No |
| 64 | CYCS | CYCS Entrez, Source | cytochrome c, somatic | 7500 | -0.010 | -0.1550 | No |
| 65 | ALS2 | ALS2 Entrez, Source | amyotrophic lateral sclerosis 2 (juvenile) | 7724 | -0.013 | -0.1701 | No |
| 66 | STAT1 | STAT1 Entrez, Source | signal transducer and activator of transcription 1, 91kDa | 7778 | -0.013 | -0.1723 | No |
| 67 | BAX | BAX Entrez, Source | BCL2-associated X protein | 7784 | -0.013 | -0.1708 | No |
| 68 | XRCC4 | XRCC4 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 4 | 7798 | -0.014 | -0.1699 | No |
| 69 | CD81 | CD81 Entrez, Source | CD81 molecule | 8145 | -0.017 | -0.1937 | No |
| 70 | APOA5 | APOA5 Entrez, Source | apolipoprotein A-V | 8409 | -0.020 | -0.2109 | No |
| 71 | EDG2 | EDG2 Entrez, Source | endothelial differentiation, lysophosphatidic acid G-protein-coupled receptor, 2 | 8571 | -0.022 | -0.2201 | No |
| 72 | HIP1 | HIP1 Entrez, Source | huntingtin interacting protein 1 | 8701 | -0.023 | -0.2267 | No |
| 73 | CASP9 | CASP9 Entrez, Source | caspase 9, apoptosis-related cysteine peptidase | 8779 | -0.024 | -0.2292 | No |
| 74 | PLCB2 | PLCB2 Entrez, Source | phospholipase C, beta 2 | 9169 | -0.028 | -0.2548 | No |
| 75 | PROK2 | PROK2 Entrez, Source | prokineticin 2 | 9205 | -0.028 | -0.2536 | No |
| 76 | MAP3K11 | MAP3K11 Entrez, Source | mitogen-activated protein kinase kinase kinase 11 | 9302 | -0.029 | -0.2568 | No |
| 77 | DIABLO | DIABLO Entrez, Source | diablo homolog (Drosophila) | 9345 | -0.030 | -0.2559 | No |
| 78 | CDKN2A | CDKN2A Entrez, Source | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 9542 | -0.032 | -0.2663 | No |
| 79 | PICK1 | PICK1 Entrez, Source | protein interacting with PRKCA 1 | 9587 | -0.032 | -0.2652 | No |
| 80 | ADCYAP1 | ADCYAP1 Entrez, Source | adenylate cyclase activating polypeptide 1 (pituitary) | 9783 | -0.035 | -0.2751 | No |
| 81 | NCSTN | NCSTN Entrez, Source | nicastrin | 10195 | -0.039 | -0.3007 | No |
| 82 | PKN1 | PKN1 Entrez, Source | protein kinase N1 | 10497 | -0.043 | -0.3175 | No |
| 83 | MTCH1 | MTCH1 Entrez, Source | mitochondrial carrier homolog 1 (C. elegans) | 10709 | -0.046 | -0.3270 | No |
| 84 | GNAQ | GNAQ Entrez, Source | guanine nucleotide binding protein (G protein), q polypeptide | 10825 | -0.048 | -0.3290 | No |
| 85 | CAP1 | CAP1 Entrez, Source | CAP, adenylate cyclase-associated protein 1 (yeast) | 11031 | -0.051 | -0.3374 | No |
| 86 | DBNL | DBNL Entrez, Source | drebrin-like | 11131 | -0.054 | -0.3375 | No |
| 87 | DAXX | DAXX Entrez, Source | death-associated protein 6 | 11474 | -0.060 | -0.3551 | No |
| 88 | RALBP1 | RALBP1 Entrez, Source | ralA binding protein 1 | 11641 | -0.063 | -0.3589 | Yes |
| 89 | PPAP2A | PPAP2A Entrez, Source | phosphatidic acid phosphatase type 2A | 11707 | -0.065 | -0.3549 | Yes |
| 90 | MAP2K3 | MAP2K3 Entrez, Source | mitogen-activated protein kinase kinase 3 | 11810 | -0.068 | -0.3533 | Yes |
| 91 | SMAD3 | SMAD3 Entrez, Source | SMAD, mothers against DPP homolog 3 (Drosophila) | 11928 | -0.071 | -0.3524 | Yes |
| 92 | SERINC2 | SERINC2 Entrez, Source | serine incorporator 2 | 11948 | -0.071 | -0.3440 | Yes |
| 93 | PSEN1 | PSEN1 Entrez, Source | presenilin 1 (Alzheimer disease 3) | 12004 | -0.073 | -0.3381 | Yes |
| 94 | APH1A | APH1A Entrez, Source | anterior pharynx defective 1 homolog A (C. elegans) | 12209 | -0.080 | -0.3425 | Yes |
| 95 | GAP43 | GAP43 Entrez, Source | growth associated protein 43 | 12439 | -0.088 | -0.3477 | Yes |
| 96 | GNAS | GNAS Entrez, Source | GNAS complex locus | 12462 | -0.089 | -0.3370 | Yes |
| 97 | GNA15 | GNA15 Entrez, Source | guanine nucleotide binding protein (G protein), alpha 15 (Gq class) | 12502 | -0.092 | -0.3273 | Yes |
| 98 | PIK3CB | PIK3CB Entrez, Source | phosphoinositide-3-kinase, catalytic, beta polypeptide | 12588 | -0.096 | -0.3205 | Yes |
| 99 | PAK1 | PAK1 Entrez, Source | p21/Cdc42/Rac1-activated kinase 1 (STE20 homolog, yeast) | 12764 | -0.108 | -0.3188 | Yes |
| 100 | F2R | F2R Entrez, Source | coagulation factor II (thrombin) receptor | 12839 | -0.114 | -0.3087 | Yes |
| 101 | MAP3K10 | MAP3K10 Entrez, Source | mitogen-activated protein kinase kinase kinase 10 | 13023 | -0.135 | -0.3039 | Yes |
| 102 | PSEN2 | PSEN2 Entrez, Source | presenilin 2 (Alzheimer disease 4) | 13032 | -0.136 | -0.2857 | Yes |
| 103 | RGN | RGN Entrez, Source | regucalcin (senescence marker protein-30) | 13176 | -0.172 | -0.2727 | Yes |
| 104 | SERINC5 | SERINC5 Entrez, Source | serine incorporator 5 | 13200 | -0.190 | -0.2483 | Yes |
| 105 | PLCE1 | PLCE1 Entrez, Source | phospholipase C, epsilon 1 | 13205 | -0.194 | -0.2218 | Yes |
| 106 | EDG5 | EDG5 Entrez, Source | endothelial differentiation, sphingolipid G-protein-coupled receptor, 5 | 13254 | -0.222 | -0.1948 | Yes |
| 107 | APAF1 | APAF1 Entrez, Source | apoptotic peptidase activating factor | 13280 | -0.257 | -0.1613 | Yes |
| 108 | AVPR1A | AVPR1A Entrez, Source | arginine vasopressin receptor 1A | 13282 | -0.262 | -0.1252 | Yes |
| 109 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 13302 | -0.295 | -0.0859 | Yes |
| 110 | ADRB2 | ADRB2 Entrez, Source | adrenergic, beta-2-, receptor, surface | 13304 | -0.298 | -0.0449 | Yes |
| 111 | ADORA2B | ADORA2B Entrez, Source | adenosine A2b receptor | 13316 | -0.346 | 0.0020 | Yes |