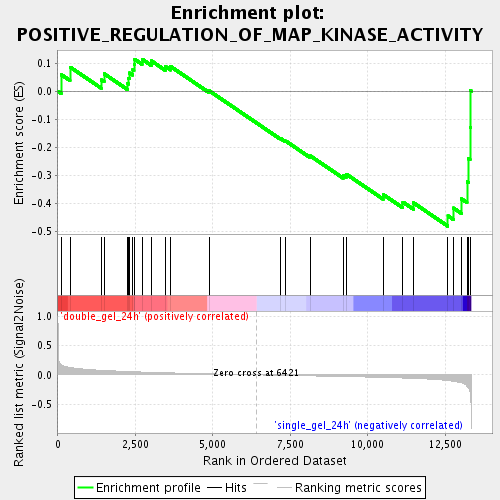
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | single_gel_24h |
| GeneSet | POSITIVE_REGULATION_OF_MAP_KINASE_ACTIVITY |
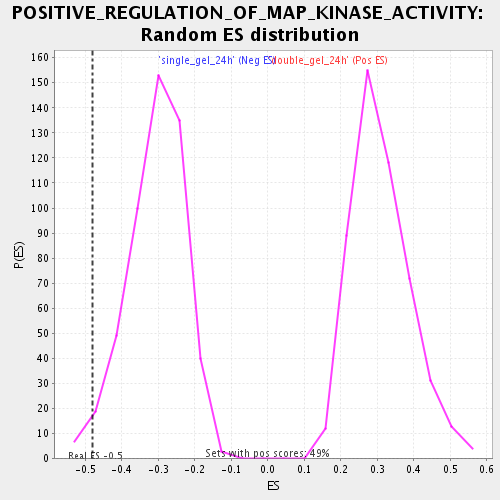
| Enrichment Score (ES) | -0.48160073 |
| Normalized Enrichment Score (NES) | -1.5708542 |
| Nominal p-value | 0.015810277 |
| FDR q-value | 0.2549674 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CHRNA7 | CHRNA7 Entrez, Source | cholinergic receptor, nicotinic, alpha 7 | 107 | 0.175 | 0.0591 | No |
| 2 | EGF | EGF Entrez, Source | epidermal growth factor (beta-urogastrone) | 392 | 0.126 | 0.0861 | No |
| 3 | ADRA2A | ADRA2A Entrez, Source | adrenergic, alpha-2A-, receptor | 1399 | 0.078 | 0.0404 | No |
| 4 | SHC1 | SHC1 Entrez, Source | SHC (Src homology 2 domain containing) transforming protein 1 | 1493 | 0.075 | 0.0622 | No |
| 5 | FGF2 | FGF2 Entrez, Source | fibroblast growth factor 2 (basic) | 2253 | 0.056 | 0.0268 | No |
| 6 | GNG3 | GNG3 Entrez, Source | guanine nucleotide binding protein (G protein), gamma 3 | 2275 | 0.056 | 0.0467 | No |
| 7 | ADRA2C | ADRA2C Entrez, Source | adrenergic, alpha-2C-, receptor | 2313 | 0.055 | 0.0651 | No |
| 8 | GHRL | GHRL Entrez, Source | ghrelin/obestatin preprohormone | 2417 | 0.053 | 0.0779 | No |
| 9 | ERBB2 | ERBB2 Entrez, Source | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | 2456 | 0.053 | 0.0952 | No |
| 10 | C5AR1 | C5AR1 Entrez, Source | complement component 5a receptor 1 | 2477 | 0.052 | 0.1137 | No |
| 11 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 2716 | 0.048 | 0.1142 | No |
| 12 | GRM4 | GRM4 Entrez, Source | glutamate receptor, metabotropic 4 | 3014 | 0.043 | 0.1083 | No |
| 13 | CD24 | CD24 Entrez, Source | CD24 molecule | 3463 | 0.036 | 0.0884 | No |
| 14 | MADD | MADD Entrez, Source | MAP-kinase activating death domain | 3637 | 0.033 | 0.0881 | No |
| 15 | TGFA | TGFA Entrez, Source | transforming growth factor, alpha | 4880 | 0.018 | 0.0016 | No |
| 16 | CXCR4 | CXCR4 Entrez, Source | chemokine (C-X-C motif) receptor 4 | 7175 | -0.007 | -0.1679 | No |
| 17 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7350 | -0.009 | -0.1775 | No |
| 18 | CD81 | CD81 Entrez, Source | CD81 molecule | 8145 | -0.017 | -0.2305 | No |
| 19 | PROK2 | PROK2 Entrez, Source | prokineticin 2 | 9205 | -0.028 | -0.2993 | No |
| 20 | MAP3K11 | MAP3K11 Entrez, Source | mitogen-activated protein kinase kinase kinase 11 | 9302 | -0.029 | -0.2953 | No |
| 21 | PKN1 | PKN1 Entrez, Source | protein kinase N1 | 10497 | -0.043 | -0.3684 | No |
| 22 | DBNL | DBNL Entrez, Source | drebrin-like | 11131 | -0.054 | -0.3954 | No |
| 23 | DAXX | DAXX Entrez, Source | death-associated protein 6 | 11474 | -0.060 | -0.3980 | No |
| 24 | PIK3CB | PIK3CB Entrez, Source | phosphoinositide-3-kinase, catalytic, beta polypeptide | 12588 | -0.096 | -0.4447 | Yes |
| 25 | PAK1 | PAK1 Entrez, Source | p21/Cdc42/Rac1-activated kinase 1 (STE20 homolog, yeast) | 12764 | -0.108 | -0.4164 | Yes |
| 26 | MAP3K10 | MAP3K10 Entrez, Source | mitogen-activated protein kinase kinase kinase 10 | 13023 | -0.135 | -0.3839 | Yes |
| 27 | PLCE1 | PLCE1 Entrez, Source | phospholipase C, epsilon 1 | 13205 | -0.194 | -0.3229 | Yes |
| 28 | EDG5 | EDG5 Entrez, Source | endothelial differentiation, sphingolipid G-protein-coupled receptor, 5 | 13254 | -0.222 | -0.2411 | Yes |
| 29 | ADRB2 | ADRB2 Entrez, Source | adrenergic, beta-2-, receptor, surface | 13304 | -0.298 | -0.1302 | Yes |
| 30 | ADORA2B | ADORA2B Entrez, Source | adenosine A2b receptor | 13316 | -0.346 | 0.0020 | Yes |