Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | single_gel_24h |
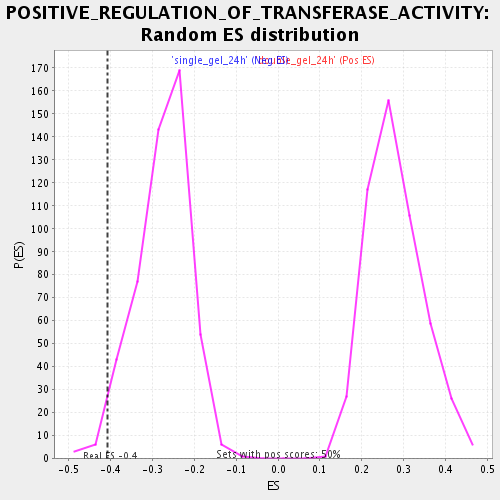
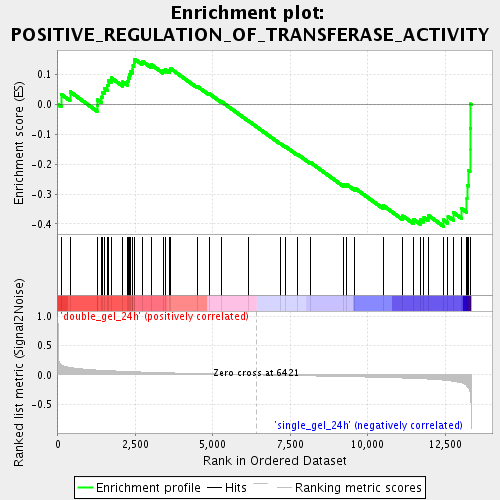
| GeneSet | POSITIVE_REGULATION_OF_TRANSFERASE_ACTIVITY |
| Enrichment Score (ES) | -0.40760955 |
| Normalized Enrichment Score (NES) | -1.4822317 |
| Nominal p-value | 0.021912351 |
| FDR q-value | 0.28277406 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CHRNA7 | CHRNA7 Entrez, Source | cholinergic receptor, nicotinic, alpha 7 | 107 | 0.175 | 0.0340 | No |
| 2 | EGF | EGF Entrez, Source | epidermal growth factor (beta-urogastrone) | 392 | 0.126 | 0.0429 | No |
| 3 | CD4 | CD4 Entrez, Source | CD4 molecule | 1266 | 0.081 | -0.0031 | No |
| 4 | CARTPT | CARTPT Entrez, Source | CART prepropeptide | 1278 | 0.081 | 0.0156 | No |
| 5 | ADRA2A | ADRA2A Entrez, Source | adrenergic, alpha-2A-, receptor | 1399 | 0.078 | 0.0252 | No |
| 6 | GADD45B | GADD45B Entrez, Source | growth arrest and DNA-damage-inducible, beta | 1431 | 0.077 | 0.0414 | No |
| 7 | SHC1 | SHC1 Entrez, Source | SHC (Src homology 2 domain containing) transforming protein 1 | 1493 | 0.075 | 0.0548 | No |
| 8 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 1596 | 0.073 | 0.0646 | No |
| 9 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 1628 | 0.072 | 0.0795 | No |
| 10 | EDN2 | EDN2 Entrez, Source | endothelin 2 | 1719 | 0.069 | 0.0894 | No |
| 11 | CHRM1 | CHRM1 Entrez, Source | cholinergic receptor, muscarinic 1 | 2092 | 0.060 | 0.0759 | No |
| 12 | FGF2 | FGF2 Entrez, Source | fibroblast growth factor 2 (basic) | 2253 | 0.056 | 0.0774 | No |
| 13 | GNG3 | GNG3 Entrez, Source | guanine nucleotide binding protein (G protein), gamma 3 | 2275 | 0.056 | 0.0893 | No |
| 14 | ADRA2C | ADRA2C Entrez, Source | adrenergic, alpha-2C-, receptor | 2313 | 0.055 | 0.0998 | No |
| 15 | TAOK2 | TAOK2 Entrez, Source | TAO kinase 2 | 2341 | 0.055 | 0.1109 | No |
| 16 | GHRL | GHRL Entrez, Source | ghrelin/obestatin preprohormone | 2417 | 0.053 | 0.1181 | No |
| 17 | PARD3 | PARD3 Entrez, Source | par-3 partitioning defective 3 homolog (C. elegans) | 2421 | 0.053 | 0.1307 | No |
| 18 | ERBB2 | ERBB2 Entrez, Source | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | 2456 | 0.053 | 0.1408 | No |
| 19 | C5AR1 | C5AR1 Entrez, Source | complement component 5a receptor 1 | 2477 | 0.052 | 0.1519 | No |
| 20 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 2716 | 0.048 | 0.1454 | No |
| 21 | GRM4 | GRM4 Entrez, Source | glutamate receptor, metabotropic 4 | 3014 | 0.043 | 0.1334 | No |
| 22 | EREG | EREG Entrez, Source | epiregulin | 3391 | 0.037 | 0.1140 | No |
| 23 | CD24 | CD24 Entrez, Source | CD24 molecule | 3463 | 0.036 | 0.1172 | No |
| 24 | PRLR | PRLR Entrez, Source | prolactin receptor | 3608 | 0.034 | 0.1145 | No |
| 25 | MADD | MADD Entrez, Source | MAP-kinase activating death domain | 3637 | 0.033 | 0.1204 | No |
| 26 | LYK5 | LYK5 Entrez, Source | - | 4491 | 0.022 | 0.0615 | No |
| 27 | TGFA | TGFA Entrez, Source | transforming growth factor, alpha | 4880 | 0.018 | 0.0366 | No |
| 28 | SERINC1 | SERINC1 Entrez, Source | serine incorporator 1 | 5271 | 0.013 | 0.0103 | No |
| 29 | CCND3 | CCND3 Entrez, Source | cyclin D3 | 6145 | 0.003 | -0.0546 | No |
| 30 | CXCR4 | CXCR4 Entrez, Source | chemokine (C-X-C motif) receptor 4 | 7175 | -0.007 | -0.1303 | No |
| 31 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7350 | -0.009 | -0.1412 | No |
| 32 | ALS2 | ALS2 Entrez, Source | amyotrophic lateral sclerosis 2 (juvenile) | 7724 | -0.013 | -0.1662 | No |
| 33 | CD81 | CD81 Entrez, Source | CD81 molecule | 8145 | -0.017 | -0.1936 | No |
| 34 | PROK2 | PROK2 Entrez, Source | prokineticin 2 | 9205 | -0.028 | -0.2666 | No |
| 35 | MAP3K11 | MAP3K11 Entrez, Source | mitogen-activated protein kinase kinase kinase 11 | 9302 | -0.029 | -0.2668 | No |
| 36 | PICK1 | PICK1 Entrez, Source | protein interacting with PRKCA 1 | 9587 | -0.032 | -0.2803 | No |
| 37 | PKN1 | PKN1 Entrez, Source | protein kinase N1 | 10497 | -0.043 | -0.3383 | No |
| 38 | DBNL | DBNL Entrez, Source | drebrin-like | 11131 | -0.054 | -0.3730 | No |
| 39 | DAXX | DAXX Entrez, Source | death-associated protein 6 | 11474 | -0.060 | -0.3843 | No |
| 40 | PPAP2A | PPAP2A Entrez, Source | phosphatidic acid phosphatase type 2A | 11707 | -0.065 | -0.3862 | No |
| 41 | MAP2K3 | MAP2K3 Entrez, Source | mitogen-activated protein kinase kinase 3 | 11810 | -0.068 | -0.3776 | No |
| 42 | SERINC2 | SERINC2 Entrez, Source | serine incorporator 2 | 11948 | -0.071 | -0.3707 | No |
| 43 | GAP43 | GAP43 Entrez, Source | growth associated protein 43 | 12439 | -0.088 | -0.3863 | Yes |
| 44 | PIK3CB | PIK3CB Entrez, Source | phosphoinositide-3-kinase, catalytic, beta polypeptide | 12588 | -0.096 | -0.3744 | Yes |
| 45 | PAK1 | PAK1 Entrez, Source | p21/Cdc42/Rac1-activated kinase 1 (STE20 homolog, yeast) | 12764 | -0.108 | -0.3616 | Yes |
| 46 | MAP3K10 | MAP3K10 Entrez, Source | mitogen-activated protein kinase kinase kinase 10 | 13023 | -0.135 | -0.3485 | Yes |
| 47 | SERINC5 | SERINC5 Entrez, Source | serine incorporator 5 | 13200 | -0.190 | -0.3160 | Yes |
| 48 | PLCE1 | PLCE1 Entrez, Source | phospholipase C, epsilon 1 | 13205 | -0.194 | -0.2696 | Yes |
| 49 | EDG5 | EDG5 Entrez, Source | endothelial differentiation, sphingolipid G-protein-coupled receptor, 5 | 13254 | -0.222 | -0.2197 | Yes |
| 50 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 13302 | -0.295 | -0.1522 | Yes |
| 51 | ADRB2 | ADRB2 Entrez, Source | adrenergic, beta-2-, receptor, surface | 13304 | -0.298 | -0.0805 | Yes |
| 52 | ADORA2B | ADORA2B Entrez, Source | adenosine A2b receptor | 13316 | -0.346 | 0.0020 | Yes |