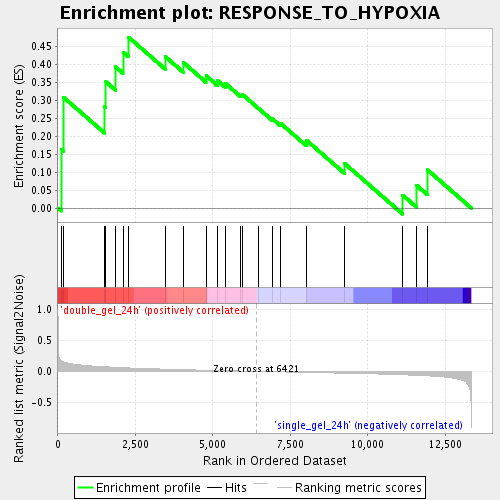
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | RESPONSE_TO_HYPOXIA |
| Enrichment Score (ES) | 0.47456974 |
| Normalized Enrichment Score (NES) | 1.4455205 |
| Nominal p-value | 0.078431375 |
| FDR q-value | 0.3015583 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CHRNA7 | CHRNA7 Entrez, Source | cholinergic receptor, nicotinic, alpha 7 | 107 | 0.175 | 0.1637 | Yes |
| 2 | CREBBP | CREBBP Entrez, Source | CREB binding protein (Rubinstein-Taybi syndrome) | 179 | 0.152 | 0.3080 | Yes |
| 3 | EPAS1 | EPAS1 Entrez, Source | endothelial PAS domain protein 1 | 1507 | 0.074 | 0.2816 | Yes |
| 4 | NF1 | NF1 Entrez, Source | neurofibromin 1 (neurofibromatosis, von Recklinghausen disease, Watson disease) | 1548 | 0.073 | 0.3508 | Yes |
| 5 | MT3 | MT3 Entrez, Source | metallothionein 3 (growth inhibitory factor (neurotrophic)) | 1858 | 0.066 | 0.3924 | Yes |
| 6 | CHRNB2 | CHRNB2 Entrez, Source | cholinergic receptor, nicotinic, beta 2 (neuronal) | 2101 | 0.060 | 0.4333 | Yes |
| 7 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 2282 | 0.056 | 0.4746 | Yes |
| 8 | CD24 | CD24 Entrez, Source | CD24 molecule | 3463 | 0.036 | 0.4211 | No |
| 9 | ARNT2 | ARNT2 Entrez, Source | aryl-hydrocarbon receptor nuclear translocator 2 | 4050 | 0.028 | 0.4043 | No |
| 10 | BNIP3 | BNIP3 Entrez, Source | BCL2/adenovirus E1B 19kDa interacting protein 3 | 4780 | 0.019 | 0.3680 | No |
| 11 | ALAS2 | ALAS2 Entrez, Source | aminolevulinate, delta-, synthase 2 (sideroblastic/hypochromic anemia) | 5149 | 0.014 | 0.3545 | No |
| 12 | PLOD1 | PLOD1 Entrez, Source | procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | 5411 | 0.011 | 0.3459 | No |
| 13 | CHRNA4 | CHRNA4 Entrez, Source | cholinergic receptor, nicotinic, alpha 4 | 5906 | 0.005 | 0.3142 | No |
| 14 | SMAD4 | SMAD4 Entrez, Source | SMAD, mothers against DPP homolog 4 (Drosophila) | 5961 | 0.005 | 0.3150 | No |
| 15 | NARFL | NARFL Entrez, Source | nuclear prelamin A recognition factor-like | 6460 | -0.000 | 0.2780 | No |
| 16 | CLDN3 | CLDN3 Entrez, Source | claudin 3 | 6913 | -0.005 | 0.2488 | No |
| 17 | CXCR4 | CXCR4 Entrez, Source | chemokine (C-X-C motif) receptor 4 | 7175 | -0.007 | 0.2363 | No |
| 18 | EGLN2 | EGLN2 Entrez, Source | egl nine homolog 2 (C. elegans) | 8020 | -0.016 | 0.1885 | No |
| 19 | EGLN1 | EGLN1 Entrez, Source | egl nine homolog 1 (C. elegans) | 9241 | -0.028 | 0.1249 | No |
| 20 | EP300 | EP300 Entrez, Source | E1A binding protein p300 | 11127 | -0.053 | 0.0360 | No |
| 21 | HIF1A | HIF1A Entrez, Source | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | 11568 | -0.062 | 0.0637 | No |
| 22 | SMAD3 | SMAD3 Entrez, Source | SMAD, mothers against DPP homolog 3 (Drosophila) | 11928 | -0.071 | 0.1061 | No |

