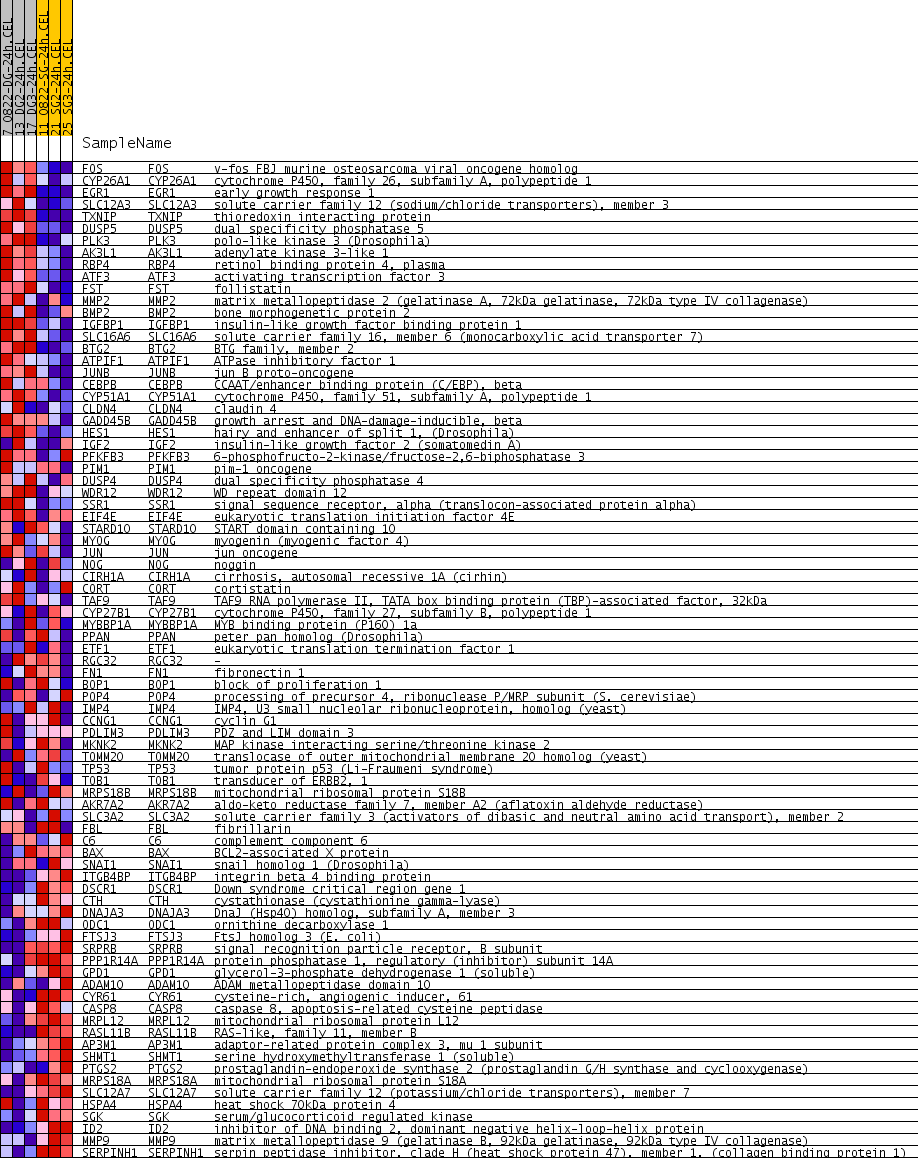
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | double_gel_24h |
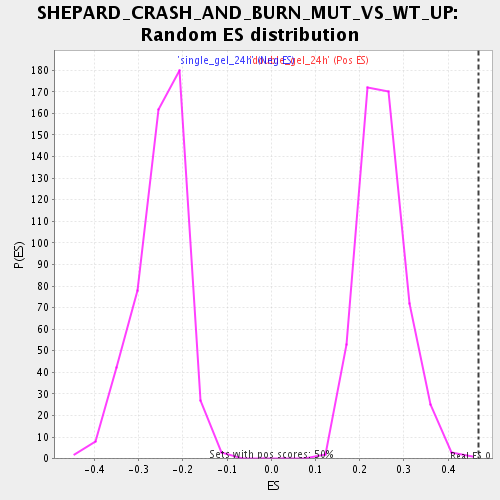
| GeneSet | SHEPARD_CRASH_AND_BURN_MUT_VS_WT_UP |
| Enrichment Score (ES) | 0.46804896 |
| Normalized Enrichment Score (NES) | 1.8660101 |
| Nominal p-value | 0.002008032 |
| FDR q-value | 0.0638106 |
| FWER p-Value | 0.862 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FOS | FOS Entrez, Source | v-fos FBJ murine osteosarcoma viral oncogene homolog | 11 | 0.331 | 0.0628 | Yes |
| 2 | CYP26A1 | CYP26A1 Entrez, Source | cytochrome P450, family 26, subfamily A, polypeptide 1 | 27 | 0.239 | 0.1076 | Yes |
| 3 | EGR1 | EGR1 Entrez, Source | early growth response 1 | 35 | 0.229 | 0.1510 | Yes |
| 4 | SLC12A3 | SLC12A3 Entrez, Source | solute carrier family 12 (sodium/chloride transporters), member 3 | 77 | 0.188 | 0.1841 | Yes |
| 5 | TXNIP | TXNIP Entrez, Source | thioredoxin interacting protein | 82 | 0.186 | 0.2194 | Yes |
| 6 | DUSP5 | DUSP5 Entrez, Source | dual specificity phosphatase 5 | 206 | 0.148 | 0.2386 | Yes |
| 7 | PLK3 | PLK3 Entrez, Source | polo-like kinase 3 (Drosophila) | 399 | 0.126 | 0.2483 | Yes |
| 8 | AK3L1 | AK3L1 Entrez, Source | adenylate kinase 3-like 1 | 418 | 0.124 | 0.2708 | Yes |
| 9 | RBP4 | RBP4 Entrez, Source | retinol binding protein 4, plasma | 487 | 0.119 | 0.2884 | Yes |
| 10 | ATF3 | ATF3 Entrez, Source | activating transcription factor 3 | 502 | 0.118 | 0.3100 | Yes |
| 11 | FST | FST Entrez, Source | follistatin | 503 | 0.117 | 0.3325 | Yes |
| 12 | MMP2 | MMP2 Entrez, Source | matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) | 527 | 0.115 | 0.3528 | Yes |
| 13 | BMP2 | BMP2 Entrez, Source | bone morphogenetic protein 2 | 547 | 0.114 | 0.3732 | Yes |
| 14 | IGFBP1 | IGFBP1 Entrez, Source | insulin-like growth factor binding protein 1 | 578 | 0.112 | 0.3924 | Yes |
| 15 | SLC16A6 | SLC16A6 Entrez, Source | solute carrier family 16, member 6 (monocarboxylic acid transporter 7) | 674 | 0.105 | 0.4054 | Yes |
| 16 | BTG2 | BTG2 Entrez, Source | BTG family, member 2 | 677 | 0.105 | 0.4255 | Yes |
| 17 | ATPIF1 | ATPIF1 Entrez, Source | ATPase inhibitory factor 1 | 741 | 0.102 | 0.4404 | Yes |
| 18 | JUNB | JUNB Entrez, Source | jun B proto-oncogene | 816 | 0.099 | 0.4538 | Yes |
| 19 | CEBPB | CEBPB Entrez, Source | CCAAT/enhancer binding protein (C/EBP), beta | 1304 | 0.080 | 0.4325 | Yes |
| 20 | CYP51A1 | CYP51A1 Entrez, Source | cytochrome P450, family 51, subfamily A, polypeptide 1 | 1345 | 0.079 | 0.4446 | Yes |
| 21 | CLDN4 | CLDN4 Entrez, Source | claudin 4 | 1370 | 0.078 | 0.4578 | Yes |
| 22 | GADD45B | GADD45B Entrez, Source | growth arrest and DNA-damage-inducible, beta | 1431 | 0.077 | 0.4680 | Yes |
| 23 | HES1 | HES1 Entrez, Source | hairy and enhancer of split 1, (Drosophila) | 1845 | 0.066 | 0.4496 | No |
| 24 | IGF2 | IGF2 Entrez, Source | insulin-like growth factor 2 (somatomedin A) | 2858 | 0.045 | 0.3820 | No |
| 25 | PFKFB3 | PFKFB3 Entrez, Source | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | 3285 | 0.039 | 0.3573 | No |
| 26 | PIM1 | PIM1 Entrez, Source | pim-1 oncogene | 3360 | 0.037 | 0.3589 | No |
| 27 | DUSP4 | DUSP4 Entrez, Source | dual specificity phosphatase 4 | 3568 | 0.034 | 0.3498 | No |
| 28 | WDR12 | WDR12 Entrez, Source | WD repeat domain 12 | 4136 | 0.026 | 0.3122 | No |
| 29 | SSR1 | SSR1 Entrez, Source | signal sequence receptor, alpha (translocon-associated protein alpha) | 4186 | 0.026 | 0.3135 | No |
| 30 | EIF4E | EIF4E Entrez, Source | eukaryotic translation initiation factor 4E | 4511 | 0.022 | 0.2932 | No |
| 31 | STARD10 | STARD10 Entrez, Source | START domain containing 10 | 4710 | 0.019 | 0.2820 | No |
| 32 | MYOG | MYOG Entrez, Source | myogenin (myogenic factor 4) | 4784 | 0.019 | 0.2801 | No |
| 33 | JUN | JUN Entrez, Source | jun oncogene | 4841 | 0.018 | 0.2793 | No |
| 34 | NOG | NOG Entrez, Source | noggin | 4995 | 0.016 | 0.2709 | No |
| 35 | CIRH1A | CIRH1A Entrez, Source | cirrhosis, autosomal recessive 1A (cirhin) | 5008 | 0.016 | 0.2730 | No |
| 36 | CORT | CORT Entrez, Source | cortistatin | 5114 | 0.015 | 0.2679 | No |
| 37 | TAF9 | TAF9 Entrez, Source | TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa | 5433 | 0.011 | 0.2460 | No |
| 38 | CYP27B1 | CYP27B1 Entrez, Source | cytochrome P450, family 27, subfamily B, polypeptide 1 | 5553 | 0.010 | 0.2389 | No |
| 39 | MYBBP1A | MYBBP1A Entrez, Source | MYB binding protein (P160) 1a | 5640 | 0.009 | 0.2341 | No |
| 40 | PPAN | PPAN Entrez, Source | peter pan homolog (Drosophila) | 5662 | 0.008 | 0.2341 | No |
| 41 | ETF1 | ETF1 Entrez, Source | eukaryotic translation termination factor 1 | 5691 | 0.008 | 0.2335 | No |
| 42 | RGC32 | RGC32 Entrez, Source | - | 5929 | 0.005 | 0.2167 | No |
| 43 | FN1 | FN1 Entrez, Source | fibronectin 1 | 6064 | 0.004 | 0.2073 | No |
| 44 | BOP1 | BOP1 Entrez, Source | block of proliferation 1 | 6211 | 0.002 | 0.1967 | No |
| 45 | POP4 | POP4 Entrez, Source | processing of precursor 4, ribonuclease P/MRP subunit (S. cerevisiae) | 6269 | 0.002 | 0.1927 | No |
| 46 | IMP4 | IMP4 Entrez, Source | IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) | 6270 | 0.002 | 0.1930 | No |
| 47 | CCNG1 | CCNG1 Entrez, Source | cyclin G1 | 6322 | 0.001 | 0.1894 | No |
| 48 | PDLIM3 | PDLIM3 Entrez, Source | PDZ and LIM domain 3 | 6622 | -0.002 | 0.1673 | No |
| 49 | MKNK2 | MKNK2 Entrez, Source | MAP kinase interacting serine/threonine kinase 2 | 6661 | -0.003 | 0.1649 | No |
| 50 | TOMM20 | TOMM20 Entrez, Source | translocase of outer mitochondrial membrane 20 homolog (yeast) | 6756 | -0.003 | 0.1585 | No |
| 51 | TP53 | TP53 Entrez, Source | tumor protein p53 (Li-Fraumeni syndrome) | 6874 | -0.004 | 0.1505 | No |
| 52 | TOB1 | TOB1 Entrez, Source | transducer of ERBB2, 1 | 6990 | -0.006 | 0.1429 | No |
| 53 | MRPS18B | MRPS18B Entrez, Source | mitochondrial ribosomal protein S18B | 7041 | -0.006 | 0.1403 | No |
| 54 | AKR7A2 | AKR7A2 Entrez, Source | aldo-keto reductase family 7, member A2 (aflatoxin aldehyde reductase) | 7115 | -0.007 | 0.1360 | No |
| 55 | SLC3A2 | SLC3A2 Entrez, Source | solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 | 7310 | -0.009 | 0.1231 | No |
| 56 | FBL | FBL Entrez, Source | fibrillarin | 7429 | -0.010 | 0.1161 | No |
| 57 | C6 | C6 Entrez, Source | complement component 6 | 7757 | -0.013 | 0.0939 | No |
| 58 | BAX | BAX Entrez, Source | BCL2-associated X protein | 7784 | -0.013 | 0.0945 | No |
| 59 | SNAI1 | SNAI1 Entrez, Source | snail homolog 1 (Drosophila) | 8517 | -0.021 | 0.0434 | No |
| 60 | ITGB4BP | ITGB4BP Entrez, Source | integrin beta 4 binding protein | 8810 | -0.024 | 0.0259 | No |
| 61 | DSCR1 | DSCR1 Entrez, Source | Down syndrome critical region gene 1 | 8894 | -0.025 | 0.0245 | No |
| 62 | CTH | CTH Entrez, Source | cystathionase (cystathionine gamma-lyase) | 9069 | -0.027 | 0.0165 | No |
| 63 | DNAJA3 | DNAJA3 Entrez, Source | DnaJ (Hsp40) homolog, subfamily A, member 3 | 9342 | -0.029 | 0.0016 | No |
| 64 | ODC1 | ODC1 Entrez, Source | ornithine decarboxylase 1 | 9554 | -0.032 | -0.0081 | No |
| 65 | FTSJ3 | FTSJ3 Entrez, Source | FtsJ homolog 3 (E. coli) | 9589 | -0.033 | -0.0044 | No |
| 66 | SRPRB | SRPRB Entrez, Source | signal recognition particle receptor, B subunit | 9742 | -0.034 | -0.0093 | No |
| 67 | PPP1R14A | PPP1R14A Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 14A | 10021 | -0.037 | -0.0231 | No |
| 68 | GPD1 | GPD1 Entrez, Source | glycerol-3-phosphate dehydrogenase 1 (soluble) | 10037 | -0.038 | -0.0170 | No |
| 69 | ADAM10 | ADAM10 Entrez, Source | ADAM metallopeptidase domain 10 | 10107 | -0.038 | -0.0148 | No |
| 70 | CYR61 | CYR61 Entrez, Source | cysteine-rich, angiogenic inducer, 61 | 10367 | -0.041 | -0.0264 | No |
| 71 | CASP8 | CASP8 Entrez, Source | caspase 8, apoptosis-related cysteine peptidase | 10368 | -0.041 | -0.0185 | No |
| 72 | MRPL12 | MRPL12 Entrez, Source | mitochondrial ribosomal protein L12 | 10545 | -0.044 | -0.0233 | No |
| 73 | RASL11B | RASL11B Entrez, Source | RAS-like, family 11, member B | 11001 | -0.051 | -0.0478 | No |
| 74 | AP3M1 | AP3M1 Entrez, Source | adaptor-related protein complex 3, mu 1 subunit | 11013 | -0.051 | -0.0388 | No |
| 75 | SHMT1 | SHMT1 Entrez, Source | serine hydroxymethyltransferase 1 (soluble) | 11060 | -0.052 | -0.0323 | No |
| 76 | PTGS2 | PTGS2 Entrez, Source | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | 12422 | -0.088 | -0.1181 | No |
| 77 | MRPS18A | MRPS18A Entrez, Source | mitochondrial ribosomal protein S18A | 12449 | -0.089 | -0.1030 | No |
| 78 | SLC12A7 | SLC12A7 Entrez, Source | solute carrier family 12 (potassium/chloride transporters), member 7 | 12586 | -0.096 | -0.0949 | No |
| 79 | HSPA4 | HSPA4 Entrez, Source | heat shock 70kDa protein 4 | 12729 | -0.105 | -0.0854 | No |
| 80 | SGK | SGK Entrez, Source | serum/glucocorticoid regulated kinase | 12799 | -0.110 | -0.0695 | No |
| 81 | ID2 | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 13084 | -0.146 | -0.0628 | No |
| 82 | MMP9 | MMP9 Entrez, Source | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) | 13151 | -0.163 | -0.0365 | No |
| 83 | SERPINH1 | SERPINH1 Entrez, Source | serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) | 13285 | -0.265 | 0.0043 | No |