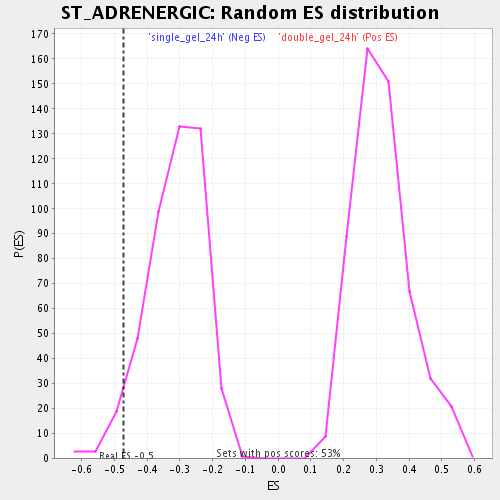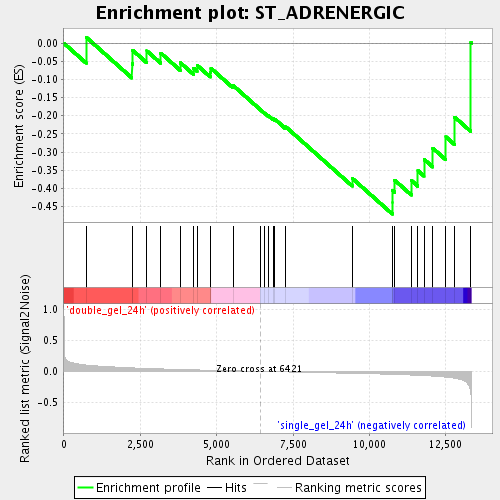
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | single_gel_24h |
| GeneSet | ST_ADRENERGIC |
| Enrichment Score (ES) | -0.4712255 |
| Normalized Enrichment Score (NES) | -1.5051818 |
| Nominal p-value | 0.04506438 |
| FDR q-value | 0.27269283 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SRC | SRC Entrez, Source | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | 729 | 0.103 | 0.0166 | No |
| 2 | PIK3R1 | PIK3R1 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 1 (p85 alpha) | 2228 | 0.057 | -0.0566 | No |
| 3 | ITPR3 | ITPR3 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 3 | 2251 | 0.056 | -0.0193 | No |
| 4 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 2716 | 0.048 | -0.0211 | No |
| 5 | KCNJ9 | KCNJ9 Entrez, Source | potassium inwardly-rectifying channel, subfamily J, member 9 | 3168 | 0.041 | -0.0269 | No |
| 6 | GNA11 | GNA11 Entrez, Source | guanine nucleotide binding protein (G protein), alpha 11 (Gq class) | 3807 | 0.031 | -0.0533 | No |
| 7 | ITPKA | ITPKA Entrez, Source | inositol 1,4,5-trisphosphate 3-kinase A | 4243 | 0.025 | -0.0684 | No |
| 8 | ITPKB | ITPKB Entrez, Source | inositol 1,4,5-trisphosphate 3-kinase B | 4358 | 0.024 | -0.0606 | No |
| 9 | KCNJ3 | KCNJ3 Entrez, Source | potassium inwardly-rectifying channel, subfamily J, member 3 | 4792 | 0.019 | -0.0802 | No |
| 10 | RAF1 | RAF1 Entrez, Source | v-raf-1 murine leukemia viral oncogene homolog 1 | 4811 | 0.018 | -0.0688 | No |
| 11 | ITPR2 | ITPR2 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 2 | 5533 | 0.010 | -0.1161 | No |
| 12 | KCNJ5 | KCNJ5 Entrez, Source | potassium inwardly-rectifying channel, subfamily J, member 5 | 6426 | -0.000 | -0.1831 | No |
| 13 | AR | AR Entrez, Source | androgen receptor (dihydrotestosterone receptor; testicular feminization; spinal and bulbar muscular atrophy; Kennedy disease) | 6572 | -0.002 | -0.1928 | No |
| 14 | MAPK14 | MAPK14 Entrez, Source | mitogen-activated protein kinase 14 | 6688 | -0.003 | -0.1994 | No |
| 15 | MAPK1 | MAPK1 Entrez, Source | mitogen-activated protein kinase 1 | 6842 | -0.004 | -0.2081 | No |
| 16 | PITX2 | PITX2 Entrez, Source | paired-like homeodomain transcription factor 2 | 6901 | -0.005 | -0.2092 | No |
| 17 | CAMP | CAMP Entrez, Source | cathelicidin antimicrobial peptide | 7261 | -0.008 | -0.2305 | No |
| 18 | PIK3CA | PIK3CA Entrez, Source | phosphoinositide-3-kinase, catalytic, alpha polypeptide | 9438 | -0.031 | -0.3727 | No |
| 19 | ITPR1 | ITPR1 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 1 | 10751 | -0.047 | -0.4386 | Yes |
| 20 | AKT1 | AKT1 Entrez, Source | v-akt murine thymoma viral oncogene homolog 1 | 10753 | -0.047 | -0.4060 | Yes |
| 21 | GNAQ | GNAQ Entrez, Source | guanine nucleotide binding protein (G protein), q polypeptide | 10825 | -0.048 | -0.3778 | Yes |
| 22 | BRAF | BRAF Entrez, Source | v-raf murine sarcoma viral oncogene homolog B1 | 11372 | -0.058 | -0.3785 | Yes |
| 23 | MAPK10 | MAPK10 Entrez, Source | mitogen-activated protein kinase 10 | 11586 | -0.062 | -0.3514 | Yes |
| 24 | DAG1 | DAG1 Entrez, Source | dystroglycan 1 (dystrophin-associated glycoprotein 1) | 11785 | -0.067 | -0.3200 | Yes |
| 25 | APC | APC Entrez, Source | adenomatosis polyposis coli | 12071 | -0.075 | -0.2894 | Yes |
| 26 | GNA15 | GNA15 Entrez, Source | guanine nucleotide binding protein (G protein), alpha 15 (Gq class) | 12502 | -0.092 | -0.2583 | Yes |
| 27 | ASAH1 | ASAH1 Entrez, Source | N-acylsphingosine amidohydrolase (acid ceramidase) 1 | 12784 | -0.110 | -0.2034 | Yes |
| 28 | GNAI1 | GNAI1 Entrez, Source | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 | 13318 | -0.354 | 0.0018 | Yes |