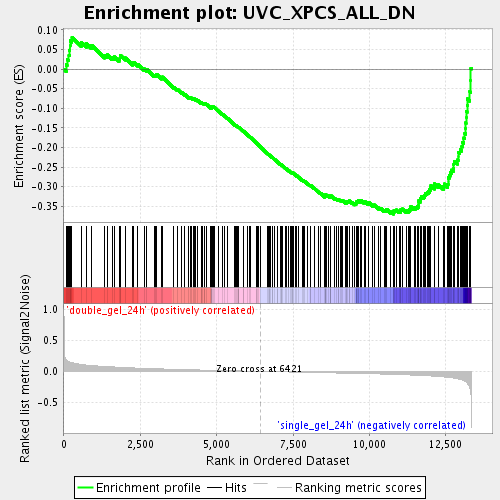
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | single_gel_24h |
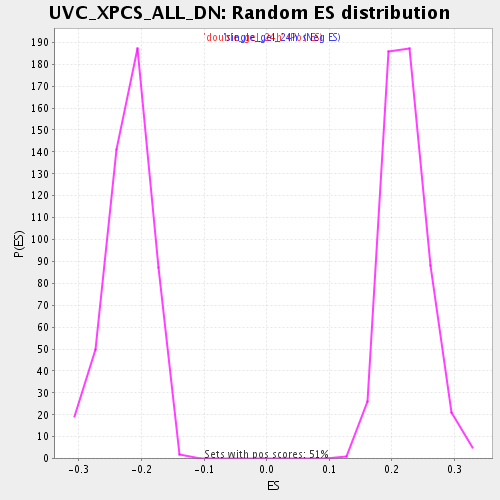
| GeneSet | UVC_XPCS_ALL_DN |
| Enrichment Score (ES) | -0.36987412 |
| Normalized Enrichment Score (NES) | -1.6792818 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.16917165 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IGF1R | IGF1R Entrez, Source | insulin-like growth factor 1 receptor | 76 | 0.189 | 0.0114 | No |
| 2 | CXCL12 | CXCL12 Entrez, Source | chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | 105 | 0.175 | 0.0251 | No |
| 3 | BHLHB2 | BHLHB2 Entrez, Source | basic helix-loop-helix domain containing, class B, 2 | 164 | 0.155 | 0.0348 | No |
| 4 | CREBBP | CREBBP Entrez, Source | CREB binding protein (Rubinstein-Taybi syndrome) | 179 | 0.152 | 0.0476 | No |
| 5 | IGF2BP3 | IGF2BP3 Entrez, Source | insulin-like growth factor 2 mRNA binding protein 3 | 199 | 0.149 | 0.0597 | No |
| 6 | DUSP5 | DUSP5 Entrez, Source | dual specificity phosphatase 5 | 206 | 0.148 | 0.0727 | No |
| 7 | IFRD1 | IFRD1 Entrez, Source | interferon-related developmental regulator 1 | 263 | 0.140 | 0.0812 | No |
| 8 | SLK | SLK Entrez, Source | STE20-like kinase (yeast) | 565 | 0.112 | 0.0685 | No |
| 9 | MID1 | MID1 Entrez, Source | midline 1 (Opitz/BBB syndrome) | 731 | 0.103 | 0.0652 | No |
| 10 | ACSL3 | ACSL3 Entrez, Source | acyl-CoA synthetase long-chain family member 3 | 906 | 0.095 | 0.0606 | No |
| 11 | CENTB2 | CENTB2 Entrez, Source | centaurin, beta 2 | 1335 | 0.079 | 0.0352 | No |
| 12 | MYC | MYC Entrez, Source | v-myc myelocytomatosis viral oncogene homolog (avian) | 1411 | 0.077 | 0.0366 | No |
| 13 | HNRPDL | HNRPDL Entrez, Source | heterogeneous nuclear ribonucleoprotein D-like | 1582 | 0.073 | 0.0302 | No |
| 14 | ZHX3 | ZHX3 Entrez, Source | zinc fingers and homeoboxes 3 | 1652 | 0.071 | 0.0314 | No |
| 15 | SERPINE1 | SERPINE1 Entrez, Source | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 1805 | 0.067 | 0.0260 | No |
| 16 | TGFBR3 | TGFBR3 Entrez, Source | transforming growth factor, beta receptor III (betaglycan, 300kDa) | 1835 | 0.067 | 0.0298 | No |
| 17 | KLF10 | KLF10 Entrez, Source | Kruppel-like factor 10 | 1837 | 0.067 | 0.0358 | No |
| 18 | RAGE | RAGE Entrez, Source | renal tumor antigen | 2008 | 0.062 | 0.0285 | No |
| 19 | FGF2 | FGF2 Entrez, Source | fibroblast growth factor 2 (basic) | 2253 | 0.056 | 0.0150 | No |
| 20 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 2282 | 0.056 | 0.0180 | No |
| 21 | PARD3 | PARD3 Entrez, Source | par-3 partitioning defective 3 homolog (C. elegans) | 2421 | 0.053 | 0.0123 | No |
| 22 | CTBP2 | CTBP2 Entrez, Source | C-terminal binding protein 2 | 2635 | 0.049 | 0.0005 | No |
| 23 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 2716 | 0.048 | -0.0012 | No |
| 24 | KLF6 | KLF6 Entrez, Source | Kruppel-like factor 6 | 2960 | 0.044 | -0.0157 | No |
| 25 | SLIT2 | SLIT2 Entrez, Source | slit homolog 2 (Drosophila) | 3006 | 0.043 | -0.0153 | No |
| 26 | IL6 | IL6 Entrez, Source | interleukin 6 (interferon, beta 2) | 3035 | 0.043 | -0.0136 | No |
| 27 | WNT5A | WNT5A Entrez, Source | wingless-type MMTV integration site family, member 5A | 3195 | 0.040 | -0.0220 | No |
| 28 | MARCH7 | MARCH7 Entrez, Source | membrane-associated ring finger (C3HC4) 7 | 3217 | 0.040 | -0.0200 | No |
| 29 | RGS7 | RGS7 Entrez, Source | regulator of G-protein signalling 7 | 3598 | 0.034 | -0.0459 | No |
| 30 | CDH2 | CDH2 Entrez, Source | cadherin 2, type 1, N-cadherin (neuronal) | 3714 | 0.032 | -0.0517 | No |
| 31 | PLCB4 | PLCB4 Entrez, Source | phospholipase C, beta 4 | 3860 | 0.030 | -0.0600 | No |
| 32 | UBE2D2 | UBE2D2 Entrez, Source | ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | 3952 | 0.029 | -0.0643 | No |
| 33 | PTPN12 | PTPN12 Entrez, Source | protein tyrosine phosphatase, non-receptor type 12 | 4086 | 0.027 | -0.0719 | No |
| 34 | GMPS | GMPS Entrez, Source | guanine monphosphate synthetase | 4135 | 0.026 | -0.0732 | No |
| 35 | MAP2K1 | MAP2K1 Entrez, Source | mitogen-activated protein kinase kinase 1 | 4172 | 0.026 | -0.0735 | No |
| 36 | CUTL1 | CUTL1 Entrez, Source | cut-like 1, CCAAT displacement protein (Drosophila) | 4235 | 0.025 | -0.0760 | No |
| 37 | PEX3 | PEX3 Entrez, Source | peroxisomal biogenesis factor 3 | 4263 | 0.025 | -0.0757 | No |
| 38 | MDN1 | MDN1 Entrez, Source | MDN1, midasin homolog (yeast) | 4316 | 0.024 | -0.0775 | No |
| 39 | E2F5 | E2F5 Entrez, Source | E2F transcription factor 5, p130-binding | 4371 | 0.024 | -0.0794 | No |
| 40 | PJA2 | PJA2 Entrez, Source | praja 2, RING-H2 motif containing | 4488 | 0.022 | -0.0863 | No |
| 41 | EIF4E | EIF4E Entrez, Source | eukaryotic translation initiation factor 4E | 4511 | 0.022 | -0.0860 | No |
| 42 | CBLB | CBLB Entrez, Source | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | 4550 | 0.021 | -0.0869 | No |
| 43 | ITSN1 | ITSN1 Entrez, Source | intersectin 1 (SH3 domain protein) | 4584 | 0.021 | -0.0876 | No |
| 44 | USP15 | USP15 Entrez, Source | ubiquitin specific peptidase 15 | 4599 | 0.021 | -0.0867 | No |
| 45 | TIAM1 | TIAM1 Entrez, Source | T-cell lymphoma invasion and metastasis 1 | 4663 | 0.020 | -0.0897 | No |
| 46 | ORC2L | ORC2L Entrez, Source | origin recognition complex, subunit 2-like (yeast) | 4798 | 0.019 | -0.0983 | No |
| 47 | DLC1 | DLC1 Entrez, Source | deleted in liver cancer 1 | 4824 | 0.018 | -0.0985 | No |
| 48 | PRKCBP1 | PRKCBP1 Entrez, Source | protein kinase C binding protein 1 | 4831 | 0.018 | -0.0973 | No |
| 49 | CREB3L2 | CREB3L2 Entrez, Source | cAMP responsive element binding protein 3-like 2 | 4834 | 0.018 | -0.0958 | No |
| 50 | EDD1 | EDD1 Entrez, Source | E3 ubiquitin protein ligase, HECT domain containing, 1 | 4878 | 0.018 | -0.0975 | No |
| 51 | PRKCA | PRKCA Entrez, Source | protein kinase C, alpha | 4883 | 0.018 | -0.0962 | No |
| 52 | XPO6 | XPO6 Entrez, Source | exportin 6 | 4917 | 0.017 | -0.0972 | No |
| 53 | PHLPP | PHLPP Entrez, Source | PH domain and leucine rich repeat protein phosphatase | 5073 | 0.015 | -0.1076 | No |
| 54 | SLC7A1 | SLC7A1 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 | 5188 | 0.014 | -0.1150 | No |
| 55 | FYN | FYN Entrez, Source | FYN oncogene related to SRC, FGR, YES | 5269 | 0.013 | -0.1199 | No |
| 56 | RAD23B | RAD23B Entrez, Source | RAD23 homolog B (S. cerevisiae) | 5366 | 0.012 | -0.1262 | No |
| 57 | HOMER1 | HOMER1 Entrez, Source | homer homolog 1 (Drosophila) | 5598 | 0.009 | -0.1429 | No |
| 58 | PRR3 | PRR3 Entrez, Source | proline rich 3 | 5620 | 0.009 | -0.1437 | No |
| 59 | RBM16 | RBM16 Entrez, Source | RNA binding motif protein 16 | 5648 | 0.009 | -0.1450 | No |
| 60 | TERF1 | TERF1 Entrez, Source | telomeric repeat binding factor (NIMA-interacting) 1 | 5667 | 0.008 | -0.1456 | No |
| 61 | TSPAN5 | TSPAN5 Entrez, Source | tetraspanin 5 | 5710 | 0.008 | -0.1481 | No |
| 62 | MYST3 | MYST3 Entrez, Source | MYST histone acetyltransferase (monocytic leukemia) 3 | 5716 | 0.008 | -0.1478 | No |
| 63 | RSN | RSN Entrez, Source | restin (Reed-Steinberg cell-expressed intermediate filament-associated protein) | 5729 | 0.008 | -0.1480 | No |
| 64 | ATRN | ATRN Entrez, Source | attractin | 5869 | 0.006 | -0.1581 | No |
| 65 | GBE1 | GBE1 Entrez, Source | glucan (1,4-alpha-), branching enzyme 1 (glycogen branching enzyme, Andersen disease, glycogen storage disease type IV) | 5875 | 0.006 | -0.1579 | No |
| 66 | ATF2 | ATF2 Entrez, Source | activating transcription factor 2 | 6021 | 0.004 | -0.1686 | No |
| 67 | PUM2 | PUM2 Entrez, Source | pumilio homolog 2 (Drosophila) | 6077 | 0.004 | -0.1724 | No |
| 68 | RBM39 | RBM39 Entrez, Source | RNA binding motif protein 39 | 6107 | 0.004 | -0.1743 | No |
| 69 | FGF5 | FGF5 Entrez, Source | fibroblast growth factor 5 | 6286 | 0.001 | -0.1877 | No |
| 70 | PAFAH1B1 | PAFAH1B1 Entrez, Source | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa | 6305 | 0.001 | -0.1890 | No |
| 71 | ADARB1 | ADARB1 Entrez, Source | adenosine deaminase, RNA-specific, B1 (RED1 homolog rat) | 6339 | 0.001 | -0.1914 | No |
| 72 | GTF2F2 | GTF2F2 Entrez, Source | general transcription factor IIF, polypeptide 2, 30kDa | 6378 | 0.000 | -0.1943 | No |
| 73 | NUMB | NUMB Entrez, Source | numb homolog (Drosophila) | 6445 | -0.000 | -0.1993 | No |
| 74 | TUBGCP3 | TUBGCP3 Entrez, Source | tubulin, gamma complex associated protein 3 | 6657 | -0.003 | -0.2151 | No |
| 75 | RAB6A | RAB6A Entrez, Source | RAB6A, member RAS oncogene family | 6696 | -0.003 | -0.2177 | No |
| 76 | ZNF148 | ZNF148 Entrez, Source | zinc finger protein 148 | 6724 | -0.003 | -0.2195 | No |
| 77 | PICALM | PICALM Entrez, Source | phosphatidylinositol binding clathrin assembly protein | 6743 | -0.003 | -0.2206 | No |
| 78 | WWP1 | WWP1 Entrez, Source | WW domain containing E3 ubiquitin protein ligase 1 | 6766 | -0.003 | -0.2220 | No |
| 79 | MYO1B | MYO1B Entrez, Source | myosin IB | 6819 | -0.004 | -0.2256 | No |
| 80 | TRIM2 | TRIM2 Entrez, Source | tripartite motif-containing 2 | 6833 | -0.004 | -0.2262 | No |
| 81 | SEPT9 | SEPT9 Entrez, Source | septin 9 | 6891 | -0.005 | -0.2301 | No |
| 82 | ZNF292 | ZNF292 Entrez, Source | zinc finger protein 292 | 6983 | -0.005 | -0.2365 | No |
| 83 | MECP2 | MECP2 Entrez, Source | methyl CpG binding protein 2 (Rett syndrome) | 6994 | -0.006 | -0.2368 | No |
| 84 | CEP170 | CEP170 Entrez, Source | centrosomal protein 170kDa | 7086 | -0.006 | -0.2431 | No |
| 85 | NUP153 | NUP153 Entrez, Source | nucleoporin 153kDa | 7107 | -0.007 | -0.2441 | No |
| 86 | AFAP | AFAP Entrez, Source | - | 7125 | -0.007 | -0.2447 | No |
| 87 | CDC2L5 | CDC2L5 Entrez, Source | cell division cycle 2-like 5 (cholinesterase-related cell division controller) | 7141 | -0.007 | -0.2452 | No |
| 88 | NFKB1 | NFKB1 Entrez, Source | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (p105) | 7260 | -0.008 | -0.2535 | No |
| 89 | NR2F2 | NR2F2 Entrez, Source | nuclear receptor subfamily 2, group F, member 2 | 7287 | -0.009 | -0.2547 | No |
| 90 | ACVR1 | ACVR1 Entrez, Source | activin A receptor, type I | 7360 | -0.009 | -0.2593 | No |
| 91 | MAP2K4 | MAP2K4 Entrez, Source | mitogen-activated protein kinase kinase 4 | 7416 | -0.010 | -0.2626 | No |
| 92 | XPO1 | XPO1 Entrez, Source | exportin 1 (CRM1 homolog, yeast) | 7439 | -0.010 | -0.2634 | No |
| 93 | R3HDM1 | R3HDM1 Entrez, Source | R3H domain containing 1 | 7477 | -0.010 | -0.2653 | No |
| 94 | ABCE1 | ABCE1 Entrez, Source | ATP-binding cassette, sub-family E (OABP), member 1 | 7480 | -0.010 | -0.2645 | No |
| 95 | CDC42BPA | CDC42BPA Entrez, Source | CDC42 binding protein kinase alpha (DMPK-like) | 7499 | -0.010 | -0.2649 | No |
| 96 | MTX2 | MTX2 Entrez, Source | metaxin 2 | 7566 | -0.011 | -0.2689 | No |
| 97 | ARIH1 | ARIH1 Entrez, Source | ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 (Drosophila) | 7627 | -0.012 | -0.2724 | No |
| 98 | ATRX | ATRX Entrez, Source | alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) | 7684 | -0.012 | -0.2756 | No |
| 99 | XRCC4 | XRCC4 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 4 | 7798 | -0.014 | -0.2830 | No |
| 100 | SON | SON Entrez, Source | SON DNA binding protein | 7838 | -0.014 | -0.2847 | No |
| 101 | ARHGEF7 | ARHGEF7 Entrez, Source | Rho guanine nucleotide exchange factor (GEF) 7 | 7877 | -0.014 | -0.2863 | No |
| 102 | THBS2 | THBS2 Entrez, Source | thrombospondin 2 | 7976 | -0.015 | -0.2923 | No |
| 103 | PCMT1 | PCMT1 Entrez, Source | protein-L-isoaspartate (D-aspartate) O-methyltransferase | 8056 | -0.016 | -0.2969 | No |
| 104 | MKLN1 | MKLN1 Entrez, Source | muskelin 1, intracellular mediator containing kelch motifs | 8067 | -0.016 | -0.2961 | No |
| 105 | STK39 | STK39 Entrez, Source | serine threonine kinase 39 (STE20/SPS1 homolog, yeast) | 8189 | -0.018 | -0.3038 | No |
| 106 | MTMR3 | MTMR3 Entrez, Source | myotubularin related protein 3 | 8338 | -0.019 | -0.3133 | No |
| 107 | NUP98 | NUP98 Entrez, Source | nucleoporin 98kDa | 8404 | -0.020 | -0.3165 | No |
| 108 | CDS2 | CDS2 Entrez, Source | CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 | 8543 | -0.021 | -0.3250 | No |
| 109 | MGLL | MGLL Entrez, Source | monoglyceride lipase | 8545 | -0.021 | -0.3232 | No |
| 110 | MARK3 | MARK3 Entrez, Source | MAP/microtubule affinity-regulating kinase 3 | 8547 | -0.021 | -0.3213 | No |
| 111 | NFYC | NFYC Entrez, Source | nuclear transcription factor Y, gamma | 8563 | -0.021 | -0.3205 | No |
| 112 | GLRB | GLRB Entrez, Source | glycine receptor, beta | 8597 | -0.022 | -0.3211 | No |
| 113 | GRK5 | GRK5 Entrez, Source | G protein-coupled receptor kinase 5 | 8649 | -0.022 | -0.3229 | No |
| 114 | ZMYND11 | ZMYND11 Entrez, Source | zinc finger, MYND domain containing 11 | 8725 | -0.023 | -0.3265 | No |
| 115 | OPA1 | OPA1 Entrez, Source | optic atrophy 1 (autosomal dominant) | 8736 | -0.023 | -0.3252 | No |
| 116 | UBE2G1 | UBE2G1 Entrez, Source | ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) | 8739 | -0.023 | -0.3232 | No |
| 117 | MYO9B | MYO9B Entrez, Source | myosin IXB | 8864 | -0.025 | -0.3304 | No |
| 118 | CLOCK | CLOCK Entrez, Source | clock homolog (mouse) | 8909 | -0.025 | -0.3315 | No |
| 119 | STAU2 | STAU2 Entrez, Source | staufen, RNA binding protein, homolog 2 (Drosophila) | 8971 | -0.026 | -0.3338 | No |
| 120 | ATP2B1 | ATP2B1 Entrez, Source | ATPase, Ca++ transporting, plasma membrane 1 | 8980 | -0.026 | -0.3321 | No |
| 121 | DYRK1A | DYRK1A Entrez, Source | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | 9041 | -0.026 | -0.3342 | No |
| 122 | NBN | NBN Entrez, Source | nibrin | 9071 | -0.027 | -0.3340 | No |
| 123 | TRIP12 | TRIP12 Entrez, Source | thyroid hormone receptor interactor 12 | 9131 | -0.027 | -0.3360 | No |
| 124 | TJP2 | TJP2 Entrez, Source | tight junction protein 2 (zona occludens 2) | 9217 | -0.028 | -0.3399 | No |
| 125 | MBNL1 | MBNL1 Entrez, Source | muscleblind-like (Drosophila) | 9251 | -0.029 | -0.3399 | No |
| 126 | PKN2 | PKN2 Entrez, Source | protein kinase N2 | 9258 | -0.029 | -0.3377 | No |
| 127 | DPYD | DPYD Entrez, Source | dihydropyrimidine dehydrogenase | 9279 | -0.029 | -0.3366 | No |
| 128 | TOP2A | TOP2A Entrez, Source | topoisomerase (DNA) II alpha 170kDa | 9337 | -0.029 | -0.3383 | No |
| 129 | CEP350 | CEP350 Entrez, Source | centrosomal protein 350kDa | 9340 | -0.029 | -0.3357 | No |
| 130 | DLG1 | DLG1 Entrez, Source | discs, large homolog 1 (Drosophila) | 9446 | -0.031 | -0.3409 | No |
| 131 | LRPPRC | LRPPRC Entrez, Source | leucine-rich PPR-motif containing | 9523 | -0.032 | -0.3439 | No |
| 132 | SMARCA2 | SMARCA2 Entrez, Source | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | 9559 | -0.032 | -0.3436 | No |
| 133 | PTPN2 | PTPN2 Entrez, Source | protein tyrosine phosphatase, non-receptor type 2 | 9568 | -0.032 | -0.3413 | No |
| 134 | CREB1 | CREB1 Entrez, Source | cAMP responsive element binding protein 1 | 9571 | -0.032 | -0.3385 | No |
| 135 | NFIB | NFIB Entrez, Source | nuclear factor I/B | 9610 | -0.033 | -0.3384 | No |
| 136 | NIPBL | NIPBL Entrez, Source | Nipped-B homolog (Drosophila) | 9627 | -0.033 | -0.3366 | No |
| 137 | OGT | OGT Entrez, Source | O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) | 9640 | -0.033 | -0.3345 | No |
| 138 | PRIM2A | PRIM2A Entrez, Source | primase, polypeptide 2A, 58kDa | 9718 | -0.034 | -0.3373 | No |
| 139 | PRMT3 | PRMT3 Entrez, Source | protein arginine methyltransferase 3 | 9734 | -0.034 | -0.3353 | No |
| 140 | NPEPPS | NPEPPS Entrez, Source | aminopeptidase puromycin sensitive | 9822 | -0.035 | -0.3387 | No |
| 141 | NCOA3 | NCOA3 Entrez, Source | nuclear receptor coactivator 3 | 9861 | -0.036 | -0.3383 | No |
| 142 | APPBP2 | APPBP2 Entrez, Source | amyloid beta precursor protein (cytoplasmic tail) binding protein 2 | 9965 | -0.037 | -0.3429 | No |
| 143 | WDR7 | WDR7 Entrez, Source | WD repeat domain 7 | 9968 | -0.037 | -0.3397 | No |
| 144 | ADAM10 | ADAM10 Entrez, Source | ADAM metallopeptidase domain 10 | 10107 | -0.038 | -0.3467 | No |
| 145 | GSK3B | GSK3B Entrez, Source | glycogen synthase kinase 3 beta | 10151 | -0.039 | -0.3464 | No |
| 146 | DOCK9 | DOCK9 Entrez, Source | dedicator of cytokinesis 9 | 10296 | -0.041 | -0.3537 | No |
| 147 | CYR61 | CYR61 Entrez, Source | cysteine-rich, angiogenic inducer, 61 | 10367 | -0.041 | -0.3553 | No |
| 148 | FARS2 | FARS2 Entrez, Source | phenylalanine-tRNA synthetase 2 (mitochondrial) | 10489 | -0.043 | -0.3605 | No |
| 149 | EIF2C2 | EIF2C2 Entrez, Source | eukaryotic translation initiation factor 2C, 2 | 10536 | -0.044 | -0.3601 | No |
| 150 | PTEN | PTEN Entrez, Source | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 10560 | -0.044 | -0.3578 | No |
| 151 | EPS15 | EPS15 Entrez, Source | epidermal growth factor receptor pathway substrate 15 | 10698 | -0.046 | -0.3640 | No |
| 152 | HERC4 | HERC4 Entrez, Source | hect domain and RLD 4 | 10776 | -0.048 | -0.3655 | Yes |
| 153 | CKAP5 | CKAP5 Entrez, Source | cytoskeleton associated protein 5 | 10786 | -0.048 | -0.3619 | Yes |
| 154 | GNAQ | GNAQ Entrez, Source | guanine nucleotide binding protein (G protein), q polypeptide | 10825 | -0.048 | -0.3604 | Yes |
| 155 | TCF12 | TCF12 Entrez, Source | transcription factor 12 (HTF4, helix-loop-helix transcription factors 4) | 10869 | -0.049 | -0.3592 | Yes |
| 156 | PLEKHC1 | PLEKHC1 Entrez, Source | pleckstrin homology domain containing, family C (with FERM domain) member 1 | 10973 | -0.051 | -0.3624 | Yes |
| 157 | STRN3 | STRN3 Entrez, Source | striatin, calmodulin binding protein 3 | 11023 | -0.051 | -0.3615 | Yes |
| 158 | PPP1R12A | PPP1R12A Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 12A | 11026 | -0.051 | -0.3570 | Yes |
| 159 | BARD1 | BARD1 Entrez, Source | BRCA1 associated RING domain 1 | 11085 | -0.053 | -0.3566 | Yes |
| 160 | CNOT2 | CNOT2 Entrez, Source | CCR4-NOT transcription complex, subunit 2 | 11214 | -0.055 | -0.3614 | Yes |
| 161 | TDRD7 | TDRD7 Entrez, Source | tudor domain containing 7 | 11269 | -0.056 | -0.3604 | Yes |
| 162 | MTF2 | MTF2 Entrez, Source | metal response element binding transcription factor 2 | 11313 | -0.057 | -0.3584 | Yes |
| 163 | PPM1B | PPM1B Entrez, Source | protein phosphatase 1B (formerly 2C), magnesium-dependent, beta isoform | 11331 | -0.058 | -0.3545 | Yes |
| 164 | RASA1 | RASA1 Entrez, Source | RAS p21 protein activator (GTPase activating protein) 1 | 11353 | -0.058 | -0.3509 | Yes |
| 165 | ZFHX1B | ZFHX1B Entrez, Source | zinc finger homeobox 1b | 11458 | -0.060 | -0.3534 | Yes |
| 166 | REV3L | REV3L Entrez, Source | REV3-like, catalytic subunit of DNA polymerase zeta (yeast) | 11512 | -0.061 | -0.3519 | Yes |
| 167 | CNOT4 | CNOT4 Entrez, Source | CCR4-NOT transcription complex, subunit 4 | 11569 | -0.062 | -0.3505 | Yes |
| 168 | ID1 | ID1 Entrez, Source | inhibitor of DNA binding 1, dominant negative helix-loop-helix protein | 11598 | -0.062 | -0.3470 | Yes |
| 169 | CDC27 | CDC27 Entrez, Source | cell division cycle 27 | 11606 | -0.063 | -0.3418 | Yes |
| 170 | PTK2 | PTK2 Entrez, Source | PTK2 protein tyrosine kinase 2 | 11607 | -0.063 | -0.3361 | Yes |
| 171 | FUT8 | FUT8 Entrez, Source | fucosyltransferase 8 (alpha (1,6) fucosyltransferase) | 11656 | -0.064 | -0.3340 | Yes |
| 172 | KPNA3 | KPNA3 Entrez, Source | karyopherin alpha 3 (importin alpha 4) | 11673 | -0.064 | -0.3294 | Yes |
| 173 | RYK | RYK Entrez, Source | RYK receptor-like tyrosine kinase | 11696 | -0.065 | -0.3252 | Yes |
| 174 | PDE8A | PDE8A Entrez, Source | phosphodiesterase 8A | 11766 | -0.066 | -0.3244 | Yes |
| 175 | UBL3 | UBL3 Entrez, Source | ubiquitin-like 3 | 11808 | -0.068 | -0.3214 | Yes |
| 176 | SLC4A7 | SLC4A7 Entrez, Source | solute carrier family 4, sodium bicarbonate cotransporter, member 7 | 11826 | -0.068 | -0.3165 | Yes |
| 177 | PCTK2 | PCTK2 Entrez, Source | PCTAIRE protein kinase 2 | 11883 | -0.070 | -0.3144 | Yes |
| 178 | SMAD3 | SMAD3 Entrez, Source | SMAD, mothers against DPP homolog 3 (Drosophila) | 11928 | -0.071 | -0.3114 | Yes |
| 179 | CLASP2 | CLASP2 Entrez, Source | cytoplasmic linker associated protein 2 | 11968 | -0.072 | -0.3078 | Yes |
| 180 | PTPRK | PTPRK Entrez, Source | protein tyrosine phosphatase, receptor type, K | 11989 | -0.073 | -0.3027 | Yes |
| 181 | BTRC | BTRC Entrez, Source | beta-transducin repeat containing | 12010 | -0.073 | -0.2976 | Yes |
| 182 | IL1RAP | IL1RAP Entrez, Source | interleukin 1 receptor accessory protein | 12123 | -0.077 | -0.2992 | Yes |
| 183 | BMPR1A | BMPR1A Entrez, Source | bone morphogenetic protein receptor, type IA | 12130 | -0.077 | -0.2926 | Yes |
| 184 | PARG | PARG Entrez, Source | poly (ADP-ribose) glycohydrolase | 12259 | -0.081 | -0.2950 | Yes |
| 185 | VEGFC | VEGFC Entrez, Source | vascular endothelial growth factor C | 12416 | -0.088 | -0.2989 | Yes |
| 186 | CASK | CASK Entrez, Source | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | 12444 | -0.088 | -0.2929 | Yes |
| 187 | PRKCI | PRKCI Entrez, Source | protein kinase C, iota | 12564 | -0.095 | -0.2934 | Yes |
| 188 | LYN | LYN Entrez, Source | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | 12577 | -0.095 | -0.2856 | Yes |
| 189 | TRIO | TRIO Entrez, Source | triple functional domain (PTPRF interacting) | 12578 | -0.095 | -0.2770 | Yes |
| 190 | RB1 | RB1 Entrez, Source | retinoblastoma 1 (including osteosarcoma) | 12607 | -0.097 | -0.2703 | Yes |
| 191 | UGCG | UGCG Entrez, Source | UDP-glucose ceramide glucosyltransferase | 12641 | -0.099 | -0.2638 | Yes |
| 192 | STK3 | STK3 Entrez, Source | serine/threonine kinase 3 (STE20 homolog, yeast) | 12679 | -0.101 | -0.2574 | Yes |
| 193 | ATP2C1 | ATP2C1 Entrez, Source | ATPase, Ca++ transporting, type 2C, member 1 | 12741 | -0.106 | -0.2524 | Yes |
| 194 | MSH2 | MSH2 Entrez, Source | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | 12758 | -0.107 | -0.2439 | Yes |
| 195 | PIK3C3 | PIK3C3 Entrez, Source | phosphoinositide-3-kinase, class 3 | 12766 | -0.108 | -0.2346 | Yes |
| 196 | PAWR | PAWR Entrez, Source | PRKC, apoptosis, WT1, regulator | 12871 | -0.118 | -0.2318 | Yes |
| 197 | PDLIM5 | PDLIM5 Entrez, Source | PDZ and LIM domain 5 | 12904 | -0.122 | -0.2231 | Yes |
| 198 | AKAP9 | AKAP9 Entrez, Source | A kinase (PRKA) anchor protein (yotiao) 9 | 12906 | -0.122 | -0.2121 | Yes |
| 199 | KIF2A | KIF2A Entrez, Source | kinesin heavy chain member 2A | 12962 | -0.127 | -0.2048 | Yes |
| 200 | NMT2 | NMT2 Entrez, Source | N-myristoyltransferase 2 | 13024 | -0.135 | -0.1971 | Yes |
| 201 | NRG1 | NRG1 Entrez, Source | neuregulin 1 | 13047 | -0.140 | -0.1861 | Yes |
| 202 | RRAS2 | RRAS2 Entrez, Source | related RAS viral (r-ras) oncogene homolog 2 | 13088 | -0.147 | -0.1758 | Yes |
| 203 | RUNX1 | RUNX1 Entrez, Source | runt-related transcription factor 1 (acute myeloid leukemia 1; aml1 oncogene) | 13119 | -0.153 | -0.1642 | Yes |
| 204 | ROCK2 | ROCK2 Entrez, Source | Rho-associated, coiled-coil containing protein kinase 2 | 13146 | -0.160 | -0.1516 | Yes |
| 205 | TLE4 | TLE4 Entrez, Source | transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) | 13153 | -0.163 | -0.1373 | Yes |
| 206 | SOS2 | SOS2 Entrez, Source | son of sevenless homolog 2 (Drosophila) | 13162 | -0.167 | -0.1227 | Yes |
| 207 | CLASP1 | CLASP1 Entrez, Source | cytoplasmic linker associated protein 1 | 13168 | -0.170 | -0.1077 | Yes |
| 208 | SCHIP1 | SCHIP1 Entrez, Source | schwannomin interacting protein 1 | 13195 | -0.185 | -0.0928 | Yes |
| 209 | PLCE1 | PLCE1 Entrez, Source | phospholipase C, epsilon 1 | 13205 | -0.194 | -0.0759 | Yes |
| 210 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 13284 | -0.263 | -0.0580 | Yes |
| 211 | F3 | F3 Entrez, Source | coagulation factor III (thromboplastin, tissue factor) | 13312 | -0.331 | -0.0300 | Yes |
| 212 | GNAI1 | GNAI1 Entrez, Source | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 | 13318 | -0.354 | 0.0018 | Yes |