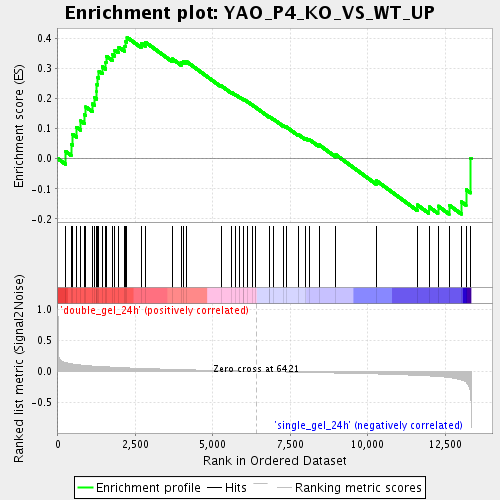
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_24h_versus_single_gel_24h.class.cls #double_gel_24h_versus_single_gel_24h_repos |
| Phenotype | class.cls#double_gel_24h_versus_single_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | YAO_P4_KO_VS_WT_UP |
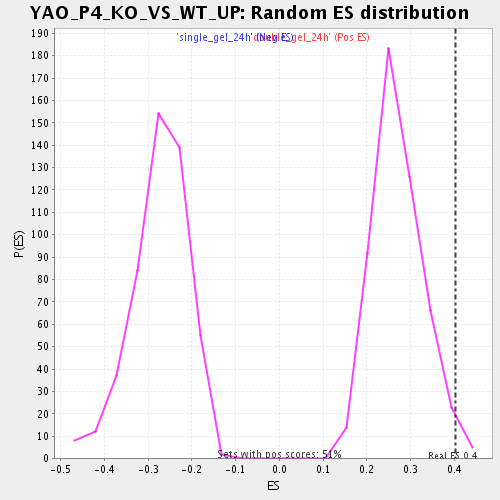
| Enrichment Score (ES) | 0.40327206 |
| Normalized Enrichment Score (NES) | 1.4917762 |
| Nominal p-value | 0.015717093 |
| FDR q-value | 0.27612048 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HMGCR | HMGCR Entrez, Source | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | 250 | 0.141 | 0.0240 | Yes |
| 2 | ENPP2 | ENPP2 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) | 429 | 0.123 | 0.0479 | Yes |
| 3 | NBL1 | NBL1 Entrez, Source | neuroblastoma, suppression of tumorigenicity 1 | 470 | 0.120 | 0.0812 | Yes |
| 4 | CYP2E1 | CYP2E1 Entrez, Source | cytochrome P450, family 2, subfamily E, polypeptide 1 | 601 | 0.110 | 0.1048 | Yes |
| 5 | CRIP2 | CRIP2 Entrez, Source | cysteine-rich protein 2 | 738 | 0.103 | 0.1257 | Yes |
| 6 | ADIPOQ | ADIPOQ Entrez, Source | adiponectin, C1Q and collagen domain containing | 852 | 0.097 | 0.1466 | Yes |
| 7 | ELN | ELN Entrez, Source | elastin (supravalvular aortic stenosis, Williams-Beuren syndrome) | 900 | 0.095 | 0.1720 | Yes |
| 8 | KCNAB1 | KCNAB1 Entrez, Source | potassium voltage-gated channel, shaker-related subfamily, beta member 1 | 1117 | 0.087 | 0.1820 | Yes |
| 9 | CFD | CFD Entrez, Source | complement factor D (adipsin) | 1187 | 0.084 | 0.2023 | Yes |
| 10 | WNT4 | WNT4 Entrez, Source | wingless-type MMTV integration site family, member 4 | 1250 | 0.082 | 0.2224 | Yes |
| 11 | DDX50 | DDX50 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 | 1256 | 0.082 | 0.2468 | Yes |
| 12 | CDKN1C | CDKN1C Entrez, Source | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 1287 | 0.081 | 0.2690 | Yes |
| 13 | REM1 | REM1 Entrez, Source | RAS (RAD and GEM)-like GTP-binding 1 | 1324 | 0.080 | 0.2904 | Yes |
| 14 | RBP1 | RBP1 Entrez, Source | retinol binding protein 1, cellular | 1436 | 0.077 | 0.3053 | Yes |
| 15 | SDPR | SDPR Entrez, Source | serum deprivation response (phosphatidylserine binding protein) | 1545 | 0.074 | 0.3195 | Yes |
| 16 | FLT1 | FLT1 Entrez, Source | fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) | 1559 | 0.073 | 0.3407 | Yes |
| 17 | CDH16 | CDH16 Entrez, Source | cadherin 16, KSP-cadherin | 1767 | 0.068 | 0.3457 | Yes |
| 18 | KLF10 | KLF10 Entrez, Source | Kruppel-like factor 10 | 1837 | 0.067 | 0.3607 | Yes |
| 19 | IGFBP3 | IGFBP3 Entrez, Source | insulin-like growth factor binding protein 3 | 1964 | 0.063 | 0.3705 | Yes |
| 20 | AKAP8L | AKAP8L Entrez, Source | A kinase (PRKA) anchor protein 8-like | 2158 | 0.059 | 0.3737 | Yes |
| 21 | ADM | ADM Entrez, Source | adrenomedullin | 2189 | 0.058 | 0.3889 | Yes |
| 22 | PIK3R1 | PIK3R1 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 1 (p85 alpha) | 2228 | 0.057 | 0.4033 | Yes |
| 23 | KLF9 | KLF9 Entrez, Source | Kruppel-like factor 9 | 2689 | 0.048 | 0.3832 | No |
| 24 | THRSP | THRSP Entrez, Source | thyroid hormone responsive (SPOT14 homolog, rat) | 2819 | 0.046 | 0.3874 | No |
| 25 | AQP1 | AQP1 Entrez, Source | aquaporin 1 (Colton blood group) | 3693 | 0.033 | 0.3316 | No |
| 26 | XDH | XDH Entrez, Source | xanthine dehydrogenase | 3980 | 0.029 | 0.3188 | No |
| 27 | NR4A1 | NR4A1 Entrez, Source | nuclear receptor subfamily 4, group A, member 1 | 4037 | 0.028 | 0.3230 | No |
| 28 | TBX3 | TBX3 Entrez, Source | T-box 3 (ulnar mammary syndrome) | 4134 | 0.027 | 0.3238 | No |
| 29 | NDRG2 | NDRG2 Entrez, Source | NDRG family member 2 | 5272 | 0.013 | 0.2421 | No |
| 30 | PDPN | PDPN Entrez, Source | podoplanin | 5603 | 0.009 | 0.2200 | No |
| 31 | THBD | THBD Entrez, Source | thrombomodulin | 5742 | 0.007 | 0.2119 | No |
| 32 | GSTT1 | GSTT1 Entrez, Source | glutathione S-transferase theta 1 | 5866 | 0.006 | 0.2044 | No |
| 33 | LCN2 | LCN2 Entrez, Source | lipocalin 2 (oncogene 24p3) | 5994 | 0.005 | 0.1963 | No |
| 34 | NCAM1 | NCAM1 Entrez, Source | neural cell adhesion molecule 1 | 5996 | 0.005 | 0.1976 | No |
| 35 | SLC27A1 | SLC27A1 Entrez, Source | solute carrier family 27 (fatty acid transporter), member 1 | 6125 | 0.003 | 0.1890 | No |
| 36 | CLDN5 | CLDN5 Entrez, Source | claudin 5 (transmembrane protein deleted in velocardiofacial syndrome) | 6273 | 0.002 | 0.1784 | No |
| 37 | RNASE4 | RNASE4 Entrez, Source | ribonuclease, RNase A family, 4 | 6380 | 0.000 | 0.1705 | No |
| 38 | BCL2 | BCL2 Entrez, Source | B-cell CLL/lymphoma 2 | 6812 | -0.004 | 0.1392 | No |
| 39 | GSTA3 | GSTA3 Entrez, Source | glutathione S-transferase A3 | 6814 | -0.004 | 0.1403 | No |
| 40 | SCD | SCD Entrez, Source | stearoyl-CoA desaturase (delta-9-desaturase) | 6967 | -0.005 | 0.1305 | No |
| 41 | APOE | APOE Entrez, Source | apolipoprotein E | 7282 | -0.008 | 0.1094 | No |
| 42 | RARRES2 | RARRES2 Entrez, Source | retinoic acid receptor responder (tazarotene induced) 2 | 7361 | -0.009 | 0.1063 | No |
| 43 | LEPR | LEPR Entrez, Source | leptin receptor | 7753 | -0.013 | 0.0809 | No |
| 44 | ATP1A2 | ATP1A2 Entrez, Source | ATPase, Na+/K+ transporting, alpha 2 (+) polypeptide | 7997 | -0.016 | 0.0673 | No |
| 45 | ACTB | ACTB Entrez, Source | actin, beta | 8105 | -0.017 | 0.0643 | No |
| 46 | CYP2F2 | 8426 | -0.020 | 0.0463 | No | ||
| 47 | ARL4A | ARL4A Entrez, Source | ADP-ribosylation factor-like 4A | 8969 | -0.026 | 0.0133 | No |
| 48 | PROS1 | PROS1 Entrez, Source | protein S (alpha) | 10274 | -0.040 | -0.0727 | No |
| 49 | ID1 | ID1 Entrez, Source | inhibitor of DNA binding 1, dominant negative helix-loop-helix protein | 11598 | -0.062 | -0.1533 | No |
| 50 | GAS6 | GAS6 Entrez, Source | growth arrest-specific 6 | 11976 | -0.072 | -0.1599 | No |
| 51 | SERTAD1 | SERTAD1 Entrez, Source | SERTA domain containing 1 | 12268 | -0.082 | -0.1570 | No |
| 52 | B3GALNT1 | B3GALNT1 Entrez, Source | beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) | 12625 | -0.098 | -0.1541 | No |
| 53 | DBP | DBP Entrez, Source | D site of albumin promoter (albumin D-box) binding protein | 13022 | -0.135 | -0.1430 | No |
| 54 | REG3A | REG3A Entrez, Source | regenerating islet-derived 3 alpha | 13173 | -0.171 | -0.1026 | No |
| 55 | CA3 | CA3 Entrez, Source | carbonic anhydrase III, muscle specific | 13324 | -0.380 | 0.0014 | No |