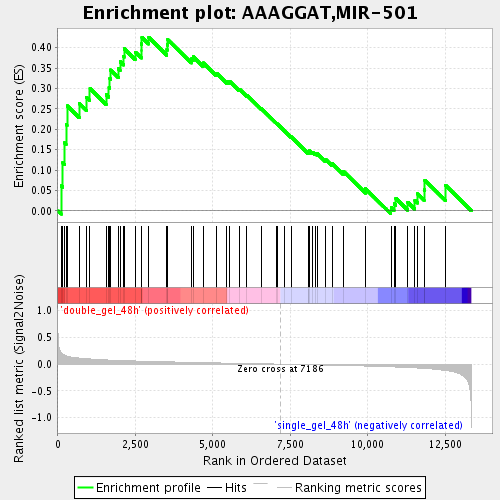
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
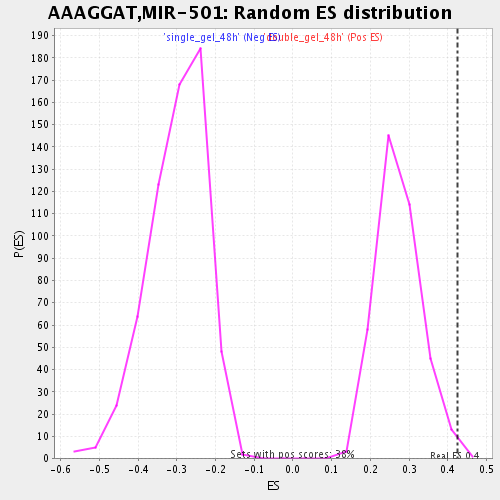
| GeneSet | AAAGGAT,MIR-501 |
| Enrichment Score (ES) | 0.42551175 |
| Normalized Enrichment Score (NES) | 1.5601679 |
| Nominal p-value | 0.007915568 |
| FDR q-value | 0.115488335 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | LPIN1 | LPIN1 Entrez, Source | lipin 1 | 104 | 0.220 | 0.0617 | Yes |
| 2 | ACACA | ACACA Entrez, Source | acetyl-Coenzyme A carboxylase alpha | 157 | 0.190 | 0.1178 | Yes |
| 3 | SRR | SRR Entrez, Source | serine racemase | 217 | 0.170 | 0.1669 | Yes |
| 4 | PITX2 | PITX2 Entrez, Source | paired-like homeodomain transcription factor 2 | 265 | 0.156 | 0.2124 | Yes |
| 5 | CART1 | CART1 Entrez, Source | cartilage paired-class homeoprotein 1 | 296 | 0.150 | 0.2575 | Yes |
| 6 | SCN3A | SCN3A Entrez, Source | sodium channel, voltage-gated, type III, alpha | 691 | 0.112 | 0.2633 | Yes |
| 7 | ADCYAP1 | ADCYAP1 Entrez, Source | adenylate cyclase activating polypeptide 1 (pituitary) | 918 | 0.101 | 0.2783 | Yes |
| 8 | TOMM70A | TOMM70A Entrez, Source | translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) | 1034 | 0.096 | 0.2999 | Yes |
| 9 | RET | RET Entrez, Source | ret proto-oncogene (multiple endocrine neoplasia and medullary thyroid carcinoma 1, Hirschsprung disease) | 1559 | 0.078 | 0.2852 | Yes |
| 10 | CUGBP2 | CUGBP2 Entrez, Source | CUG triplet repeat, RNA binding protein 2 | 1647 | 0.076 | 0.3025 | Yes |
| 11 | KCND2 | KCND2 Entrez, Source | potassium voltage-gated channel, Shal-related subfamily, member 2 | 1676 | 0.075 | 0.3241 | Yes |
| 12 | SGPP1 | SGPP1 Entrez, Source | sphingosine-1-phosphate phosphatase 1 | 1703 | 0.074 | 0.3455 | Yes |
| 13 | NR2F2 | NR2F2 Entrez, Source | nuclear receptor subfamily 2, group F, member 2 | 1957 | 0.069 | 0.3483 | Yes |
| 14 | PRKCE | PRKCE Entrez, Source | protein kinase C, epsilon | 2008 | 0.068 | 0.3659 | Yes |
| 15 | HAS2 | HAS2 Entrez, Source | hyaluronan synthase 2 | 2122 | 0.065 | 0.3781 | Yes |
| 16 | GIF | GIF Entrez, Source | gastric intrinsic factor (vitamin B synthesis) | 2137 | 0.065 | 0.3976 | Yes |
| 17 | ROBO2 | ROBO2 Entrez, Source | roundabout, axon guidance receptor, homolog 2 (Drosophila) | 2495 | 0.059 | 0.3892 | Yes |
| 18 | NFIL3 | NFIL3 Entrez, Source | nuclear factor, interleukin 3 regulated | 2690 | 0.055 | 0.3920 | Yes |
| 19 | TNNI2 | TNNI2 Entrez, Source | troponin I type 2 (skeletal, fast) | 2692 | 0.055 | 0.4094 | Yes |
| 20 | ERRFI1 | ERRFI1 Entrez, Source | ERBB receptor feedback inhibitor 1 | 2710 | 0.055 | 0.4255 | Yes |
| 21 | HES5 | HES5 Entrez, Source | hairy and enhancer of split 5 (Drosophila) | 2932 | 0.051 | 0.4251 | No |
| 22 | CD164 | CD164 Entrez, Source | CD164 molecule, sialomucin | 3516 | 0.043 | 0.3947 | No |
| 23 | ACADSB | ACADSB Entrez, Source | acyl-Coenzyme A dehydrogenase, short/branched chain | 3526 | 0.043 | 0.4074 | No |
| 24 | NPR3 | NPR3 Entrez, Source | natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) | 3545 | 0.042 | 0.4194 | No |
| 25 | TRIM39 | TRIM39 Entrez, Source | tripartite motif-containing 39 | 4304 | 0.033 | 0.3726 | No |
| 26 | CELSR2 | CELSR2 Entrez, Source | cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila) | 4367 | 0.032 | 0.3780 | No |
| 27 | UBAP1 | UBAP1 Entrez, Source | ubiquitin associated protein 1 | 4687 | 0.028 | 0.3627 | No |
| 28 | PPP1CB | PPP1CB Entrez, Source | protein phosphatase 1, catalytic subunit, beta isoform | 5113 | 0.022 | 0.3378 | No |
| 29 | VDAC2 | VDAC2 Entrez, Source | voltage-dependent anion channel 2 | 5456 | 0.019 | 0.3180 | No |
| 30 | ALS2 | ALS2 Entrez, Source | amyotrophic lateral sclerosis 2 (juvenile) | 5536 | 0.018 | 0.3177 | No |
| 31 | SNAP29 | SNAP29 Entrez, Source | synaptosomal-associated protein, 29kDa | 5854 | 0.014 | 0.2984 | No |
| 32 | SEPHS1 | SEPHS1 Entrez, Source | selenophosphate synthetase 1 | 6100 | 0.012 | 0.2837 | No |
| 33 | ATP6V1H | ATP6V1H Entrez, Source | ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H | 6568 | 0.007 | 0.2507 | No |
| 34 | CLK1 | CLK1 Entrez, Source | CDC-like kinase 1 | 7055 | 0.001 | 0.2146 | No |
| 35 | ADIPOR2 | ADIPOR2 Entrez, Source | adiponectin receptor 2 | 7084 | 0.001 | 0.2128 | No |
| 36 | SOX11 | SOX11 Entrez, Source | SRY (sex determining region Y)-box 11 | 7315 | -0.002 | 0.1960 | No |
| 37 | DCX | DCX Entrez, Source | doublecortex; lissencephaly, X-linked (doublecortin) | 7525 | -0.004 | 0.1816 | No |
| 38 | JUN | JUN Entrez, Source | jun oncogene | 8081 | -0.010 | 0.1431 | No |
| 39 | CTDSP1 | CTDSP1 Entrez, Source | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 | 8099 | -0.011 | 0.1451 | No |
| 40 | MAP3K8 | MAP3K8 Entrez, Source | mitogen-activated protein kinase kinase kinase 8 | 8105 | -0.011 | 0.1481 | No |
| 41 | MAP2K1 | MAP2K1 Entrez, Source | mitogen-activated protein kinase kinase 1 | 8219 | -0.012 | 0.1434 | No |
| 42 | PDK1 | PDK1 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 1 | 8304 | -0.013 | 0.1412 | No |
| 43 | SFRS2 | SFRS2 Entrez, Source | splicing factor, arginine/serine-rich 2 | 8369 | -0.014 | 0.1408 | No |
| 44 | NFIX | NFIX Entrez, Source | nuclear factor I/X (CCAAT-binding transcription factor) | 8634 | -0.017 | 0.1264 | No |
| 45 | QKI | QKI Entrez, Source | quaking homolog, KH domain RNA binding (mouse) | 8852 | -0.020 | 0.1163 | No |
| 46 | RDX | RDX Entrez, Source | radixin | 9216 | -0.025 | 0.0967 | No |
| 47 | SENP3 | SENP3 Entrez, Source | SUMO1/sentrin/SMT3 specific peptidase 3 | 9926 | -0.035 | 0.0545 | No |
| 48 | SLC25A3 | SLC25A3 Entrez, Source | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 | 10750 | -0.051 | 0.0085 | No |
| 49 | BCLAF1 | BCLAF1 Entrez, Source | BCL2-associated transcription factor 1 | 10849 | -0.053 | 0.0178 | No |
| 50 | NR4A3 | NR4A3 Entrez, Source | nuclear receptor subfamily 4, group A, member 3 | 10905 | -0.054 | 0.0306 | No |
| 51 | LEPROTL1 | LEPROTL1 Entrez, Source | leptin receptor overlapping transcript-like 1 | 11290 | -0.062 | 0.0213 | No |
| 52 | SYNJ1 | SYNJ1 Entrez, Source | synaptojanin 1 | 11520 | -0.069 | 0.0258 | No |
| 53 | SYT7 | SYT7 Entrez, Source | synaptotagmin VII | 11610 | -0.072 | 0.0420 | No |
| 54 | TAF5L | TAF5L Entrez, Source | TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa | 11820 | -0.080 | 0.0516 | No |
| 55 | H2AFX | H2AFX Entrez, Source | H2A histone family, member X | 11841 | -0.081 | 0.0756 | No |
| 56 | NFASC | NFASC Entrez, Source | neurofascin homolog (chicken) | 12512 | -0.118 | 0.0625 | No |