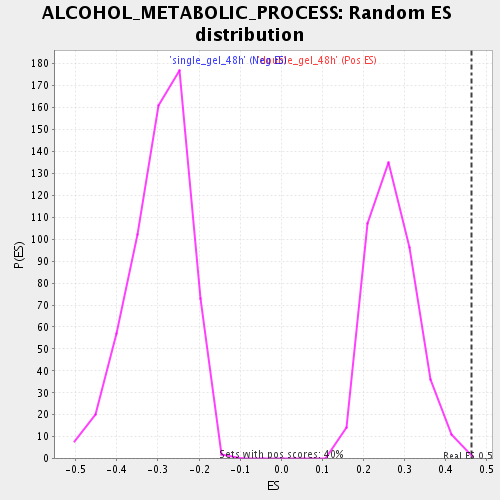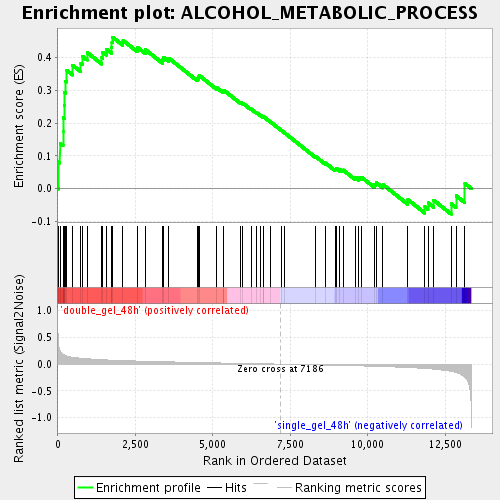
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | ALCOHOL_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.46207932 |
| Normalized Enrichment Score (NES) | 1.7143668 |
| Nominal p-value | 0.0025 |
| FDR q-value | 0.0539023 |
| FWER p-Value | 0.996 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ADH7 | ADH7 Entrez, Source | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | 19 | 0.354 | 0.0815 | Yes |
| 2 | FBP1 | FBP1 Entrez, Source | fructose-1,6-bisphosphatase 1 | 67 | 0.255 | 0.1378 | Yes |
| 3 | DHCR24 | DHCR24 Entrez, Source | 24-dehydrocholesterol reductase | 167 | 0.185 | 0.1737 | Yes |
| 4 | FDPS | FDPS Entrez, Source | farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) | 171 | 0.184 | 0.2165 | Yes |
| 5 | PDK4 | PDK4 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 4 | 207 | 0.172 | 0.2541 | Yes |
| 6 | GCK | GCK Entrez, Source | glucokinase (hexokinase 4, maturity onset diabetes of the young 2) | 218 | 0.170 | 0.2930 | Yes |
| 7 | DHCR7 | DHCR7 Entrez, Source | 7-dehydrocholesterol reductase | 247 | 0.160 | 0.3284 | Yes |
| 8 | MIOX | MIOX Entrez, Source | myo-inositol oxygenase | 289 | 0.151 | 0.3607 | Yes |
| 9 | PFKFB1 | PFKFB1 Entrez, Source | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | 476 | 0.127 | 0.3765 | Yes |
| 10 | IL4 | IL4 Entrez, Source | interleukin 4 | 741 | 0.109 | 0.3823 | Yes |
| 11 | SLC25A10 | SLC25A10 Entrez, Source | solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 | 790 | 0.107 | 0.4038 | Yes |
| 12 | ALDOB | ALDOB Entrez, Source | aldolase B, fructose-bisphosphate | 941 | 0.100 | 0.4159 | Yes |
| 13 | SULT1B1 | SULT1B1 Entrez, Source | sulfotransferase family, cytosolic, 1B, member 1 | 1416 | 0.082 | 0.3994 | Yes |
| 14 | ADH6 | ADH6 Entrez, Source | alcohol dehydrogenase 6 (class V) | 1443 | 0.081 | 0.4165 | Yes |
| 15 | UGDH | UGDH Entrez, Source | UDP-glucose dehydrogenase | 1562 | 0.078 | 0.4259 | Yes |
| 16 | NR0B2 | NR0B2 Entrez, Source | nuclear receptor subfamily 0, group B, member 2 | 1732 | 0.074 | 0.4304 | Yes |
| 17 | ACN9 | ACN9 Entrez, Source | ACN9 homolog (S. cerevisiae) | 1743 | 0.073 | 0.4468 | Yes |
| 18 | NSDHL | NSDHL Entrez, Source | NAD(P) dependent steroid dehydrogenase-like | 1768 | 0.073 | 0.4621 | Yes |
| 19 | AKR1D1 | AKR1D1 Entrez, Source | aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) | 2098 | 0.066 | 0.4528 | No |
| 20 | PPARD | PPARD Entrez, Source | peroxisome proliferative activated receptor, delta | 2558 | 0.058 | 0.4317 | No |
| 21 | GMDS | GMDS Entrez, Source | GDP-mannose 4,6-dehydratase | 2826 | 0.053 | 0.4240 | No |
| 22 | FPGT | FPGT Entrez, Source | fucose-1-phosphate guanylyltransferase | 3376 | 0.045 | 0.3932 | No |
| 23 | GFPT1 | GFPT1 Entrez, Source | glutamine-fructose-6-phosphate transaminase 1 | 3414 | 0.044 | 0.4007 | No |
| 24 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 3579 | 0.042 | 0.3982 | No |
| 25 | GFPT2 | GFPT2 Entrez, Source | glutamine-fructose-6-phosphate transaminase 2 | 4497 | 0.030 | 0.3362 | No |
| 26 | FBP2 | FBP2 Entrez, Source | fructose-1,6-bisphosphatase 2 | 4526 | 0.030 | 0.3411 | No |
| 27 | PTEN | PTEN Entrez, Source | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 4572 | 0.029 | 0.3445 | No |
| 28 | HDLBP | HDLBP Entrez, Source | high density lipoprotein binding protein (vigilin) | 5124 | 0.022 | 0.3083 | No |
| 29 | CNBP | CNBP Entrez, Source | CCHC-type zinc finger, nucleic acid binding protein | 5336 | 0.020 | 0.2971 | No |
| 30 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 5353 | 0.020 | 0.3005 | No |
| 31 | HK1 | HK1 Entrez, Source | hexokinase 1 | 5904 | 0.014 | 0.2623 | No |
| 32 | MBTPS2 | MBTPS2 Entrez, Source | membrane-bound transcription factor peptidase, site 2 | 5963 | 0.013 | 0.2610 | No |
| 33 | ADH4 | ADH4 Entrez, Source | alcohol dehydrogenase 4 (class II), pi polypeptide | 6235 | 0.011 | 0.2431 | No |
| 34 | ALDOC | ALDOC Entrez, Source | aldolase C, fructose-bisphosphate | 6404 | 0.008 | 0.2324 | No |
| 35 | PPARGC1A | PPARGC1A Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, alpha | 6549 | 0.007 | 0.2232 | No |
| 36 | IPPK | IPPK Entrez, Source | inositol 1,3,4,5,6-pentakisphosphate 2-kinase | 6623 | 0.006 | 0.2192 | No |
| 37 | GAPDHS | GAPDHS Entrez, Source | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | 6630 | 0.006 | 0.2202 | No |
| 38 | AKR1A1 | AKR1A1 Entrez, Source | aldo-keto reductase family 1, member A1 (aldehyde reductase) | 6869 | 0.003 | 0.2031 | No |
| 39 | COQ3 | COQ3 Entrez, Source | coenzyme Q3 homolog, methyltransferase (S. cerevisiae) | 7225 | -0.001 | 0.1765 | No |
| 40 | PDK2 | PDK2 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 2 | 7313 | -0.002 | 0.1703 | No |
| 41 | PDK1 | PDK1 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 1 | 8304 | -0.013 | 0.0988 | No |
| 42 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 8630 | -0.017 | 0.0784 | No |
| 43 | UGP2 | UGP2 Entrez, Source | UDP-glucose pyrophosphorylase 2 | 8944 | -0.021 | 0.0598 | No |
| 44 | ARPP-19 | ARPP-19 Entrez, Source | - | 8985 | -0.022 | 0.0619 | No |
| 45 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 9099 | -0.023 | 0.0588 | No |
| 46 | CLN8 | CLN8 Entrez, Source | ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation) | 9201 | -0.024 | 0.0569 | No |
| 47 | CEL | CEL Entrez, Source | carboxyl ester lipase (bile salt-stimulated lipase) | 9603 | -0.031 | 0.0339 | No |
| 48 | SCARB1 | SCARB1 Entrez, Source | scavenger receptor class B, member 1 | 9712 | -0.032 | 0.0333 | No |
| 49 | GALK1 | GALK1 Entrez, Source | galactokinase 1 | 9799 | -0.033 | 0.0347 | No |
| 50 | SPHK1 | SPHK1 Entrez, Source | sphingosine kinase 1 | 10209 | -0.040 | 0.0133 | No |
| 51 | CHST1 | CHST1 Entrez, Source | carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | 10266 | -0.041 | 0.0186 | No |
| 52 | GALT | GALT Entrez, Source | galactose-1-phosphate uridylyltransferase | 10491 | -0.045 | 0.0123 | No |
| 53 | PFKM | PFKM Entrez, Source | phosphofructokinase, muscle | 11295 | -0.062 | -0.0336 | No |
| 54 | HPRT1 | HPRT1 Entrez, Source | hypoxanthine phosphoribosyltransferase 1 (Lesch-Nyhan syndrome) | 11843 | -0.081 | -0.0558 | No |
| 55 | ALDOA | ALDOA Entrez, Source | aldolase A, fructose-bisphosphate | 11945 | -0.085 | -0.0435 | No |
| 56 | PFKL | PFKL Entrez, Source | phosphofructokinase, liver | 12126 | -0.093 | -0.0353 | No |
| 57 | SOAT1 | SOAT1 Entrez, Source | sterol O-acyltransferase (acyl-Coenzyme A: cholesterol acyltransferase) 1 | 12692 | -0.134 | -0.0465 | No |
| 58 | ALDH3B1 | ALDH3B1 Entrez, Source | aldehyde dehydrogenase 3 family, member B1 | 12850 | -0.156 | -0.0217 | No |
| 59 | ABCG1 | ABCG1 Entrez, Source | ATP-binding cassette, sub-family G (WHITE), member 1 | 13136 | -0.250 | 0.0155 | No |