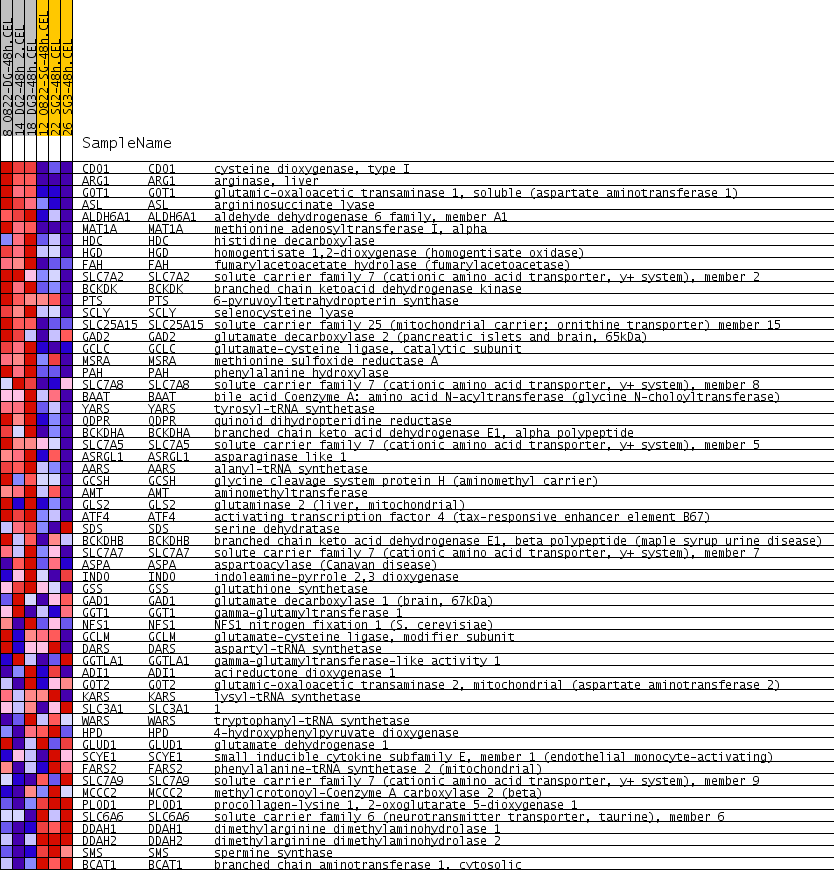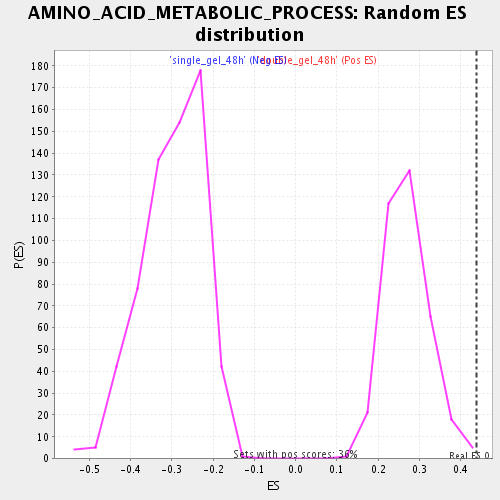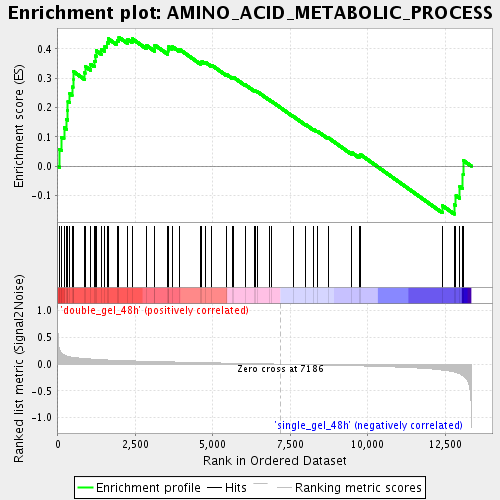
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | AMINO_ACID_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.43973505 |
| Normalized Enrichment Score (NES) | 1.6298302 |
| Nominal p-value | 0.0055710305 |
| FDR q-value | 0.085499525 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CDO1 | CDO1 Entrez, Source | cysteine dioxygenase, type I | 54 | 0.279 | 0.0576 | Yes |
| 2 | ARG1 | ARG1 Entrez, Source | arginase, liver | 117 | 0.209 | 0.0990 | Yes |
| 3 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 197 | 0.173 | 0.1314 | Yes |
| 4 | ASL | ASL Entrez, Source | argininosuccinate lyase | 273 | 0.154 | 0.1598 | Yes |
| 5 | ALDH6A1 | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 314 | 0.148 | 0.1895 | Yes |
| 6 | MAT1A | MAT1A Entrez, Source | methionine adenosyltransferase I, alpha | 323 | 0.146 | 0.2212 | Yes |
| 7 | HDC | HDC Entrez, Source | histidine decarboxylase | 383 | 0.138 | 0.2472 | Yes |
| 8 | HGD | HGD Entrez, Source | homogentisate 1,2-dioxygenase (homogentisate oxidase) | 457 | 0.129 | 0.2703 | Yes |
| 9 | FAH | FAH Entrez, Source | fumarylacetoacetate hydrolase (fumarylacetoacetase) | 489 | 0.126 | 0.2958 | Yes |
| 10 | SLC7A2 | SLC7A2 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | 494 | 0.125 | 0.3232 | Yes |
| 11 | BCKDK | BCKDK Entrez, Source | branched chain ketoacid dehydrogenase kinase | 866 | 0.104 | 0.3183 | Yes |
| 12 | PTS | PTS Entrez, Source | 6-pyruvoyltetrahydropterin synthase | 877 | 0.104 | 0.3405 | Yes |
| 13 | SCLY | SCLY Entrez, Source | selenocysteine lyase | 1056 | 0.095 | 0.3481 | Yes |
| 14 | SLC25A15 | SLC25A15 Entrez, Source | solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 | 1184 | 0.090 | 0.3584 | Yes |
| 15 | GAD2 | GAD2 Entrez, Source | glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) | 1212 | 0.089 | 0.3761 | Yes |
| 16 | GCLC | GCLC Entrez, Source | glutamate-cysteine ligase, catalytic subunit | 1237 | 0.088 | 0.3938 | Yes |
| 17 | MSRA | MSRA Entrez, Source | methionine sulfoxide reductase A | 1404 | 0.082 | 0.3996 | Yes |
| 18 | PAH | PAH Entrez, Source | phenylalanine hydroxylase | 1513 | 0.079 | 0.4089 | Yes |
| 19 | SLC7A8 | SLC7A8 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 | 1584 | 0.077 | 0.4207 | Yes |
| 20 | BAAT | BAAT Entrez, Source | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 1625 | 0.077 | 0.4347 | Yes |
| 21 | YARS | YARS Entrez, Source | tyrosyl-tRNA synthetase | 1907 | 0.070 | 0.4289 | Yes |
| 22 | QDPR | QDPR Entrez, Source | quinoid dihydropteridine reductase | 1967 | 0.069 | 0.4397 | Yes |
| 23 | BCKDHA | BCKDHA Entrez, Source | branched chain keto acid dehydrogenase E1, alpha polypeptide | 2252 | 0.063 | 0.4323 | No |
| 24 | SLC7A5 | SLC7A5 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 2396 | 0.060 | 0.4348 | No |
| 25 | ASRGL1 | ASRGL1 Entrez, Source | asparaginase like 1 | 2861 | 0.052 | 0.4115 | No |
| 26 | AARS | AARS Entrez, Source | alanyl-tRNA synthetase | 3123 | 0.048 | 0.4025 | No |
| 27 | GCSH | GCSH Entrez, Source | glycine cleavage system protein H (aminomethyl carrier) | 3126 | 0.048 | 0.4130 | No |
| 28 | AMT | AMT Entrez, Source | aminomethyltransferase | 3552 | 0.042 | 0.3904 | No |
| 29 | GLS2 | GLS2 Entrez, Source | glutaminase 2 (liver, mitochondrial) | 3555 | 0.042 | 0.3995 | No |
| 30 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 3579 | 0.042 | 0.4071 | No |
| 31 | SDS | SDS Entrez, Source | serine dehydratase | 3684 | 0.041 | 0.4082 | No |
| 32 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 3912 | 0.038 | 0.3995 | No |
| 33 | SLC7A7 | SLC7A7 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 | 4600 | 0.029 | 0.3541 | No |
| 34 | ASPA | ASPA Entrez, Source | aspartoacylase (Canavan disease) | 4645 | 0.028 | 0.3570 | No |
| 35 | INDO | INDO Entrez, Source | indoleamine-pyrrole 2,3 dioxygenase | 4751 | 0.027 | 0.3551 | No |
| 36 | GSS | GSS Entrez, Source | glutathione synthetase | 4963 | 0.024 | 0.3445 | No |
| 37 | GAD1 | GAD1 Entrez, Source | glutamate decarboxylase 1 (brain, 67kDa) | 5455 | 0.019 | 0.3117 | No |
| 38 | GGT1 | GGT1 Entrez, Source | gamma-glutamyltransferase 1 | 5638 | 0.017 | 0.3017 | No |
| 39 | NFS1 | NFS1 Entrez, Source | NFS1 nitrogen fixation 1 (S. cerevisiae) | 5674 | 0.016 | 0.3027 | No |
| 40 | GCLM | GCLM Entrez, Source | glutamate-cysteine ligase, modifier subunit | 6039 | 0.012 | 0.2780 | No |
| 41 | DARS | DARS Entrez, Source | aspartyl-tRNA synthetase | 6355 | 0.009 | 0.2563 | No |
| 42 | GGTLA1 | GGTLA1 Entrez, Source | gamma-glutamyltransferase-like activity 1 | 6376 | 0.009 | 0.2567 | No |
| 43 | ADI1 | ADI1 Entrez, Source | acireductone dioxygenase 1 | 6429 | 0.008 | 0.2547 | No |
| 44 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 6812 | 0.004 | 0.2268 | No |
| 45 | KARS | KARS Entrez, Source | lysyl-tRNA synthetase | 6885 | 0.003 | 0.2221 | No |
| 46 | SLC3A1 | SLC3A1 Entrez, Source | 1 | 7608 | -0.005 | 0.1688 | No |
| 47 | WARS | WARS Entrez, Source | tryptophanyl-tRNA synthetase | 7995 | -0.009 | 0.1418 | No |
| 48 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 8248 | -0.012 | 0.1256 | No |
| 49 | GLUD1 | GLUD1 Entrez, Source | glutamate dehydrogenase 1 | 8367 | -0.014 | 0.1198 | No |
| 50 | SCYE1 | SCYE1 Entrez, Source | small inducible cytokine subfamily E, member 1 (endothelial monocyte-activating) | 8723 | -0.018 | 0.0971 | No |
| 51 | FARS2 | FARS2 Entrez, Source | phenylalanine-tRNA synthetase 2 (mitochondrial) | 9474 | -0.029 | 0.0469 | No |
| 52 | SLC7A9 | SLC7A9 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 | 9724 | -0.032 | 0.0353 | No |
| 53 | MCCC2 | MCCC2 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 2 (beta) | 9753 | -0.033 | 0.0405 | No |
| 54 | PLOD1 | PLOD1 Entrez, Source | procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | 12413 | -0.112 | -0.1349 | No |
| 55 | SLC6A6 | SLC6A6 Entrez, Source | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | 12793 | -0.147 | -0.1310 | No |
| 56 | DDAH1 | DDAH1 Entrez, Source | dimethylarginine dimethylaminohydrolase 1 | 12847 | -0.155 | -0.1007 | No |
| 57 | DDAH2 | DDAH2 Entrez, Source | dimethylarginine dimethylaminohydrolase 2 | 12970 | -0.181 | -0.0700 | No |
| 58 | SMS | SMS Entrez, Source | spermine synthase | 13073 | -0.219 | -0.0292 | No |
| 59 | BCAT1 | BCAT1 Entrez, Source | branched chain aminotransferase 1, cytosolic | 13084 | -0.224 | 0.0194 | No |