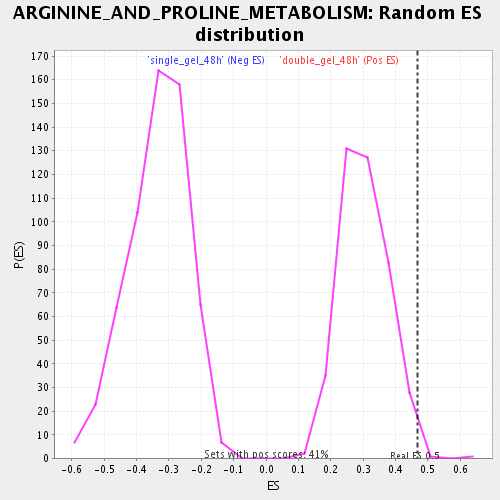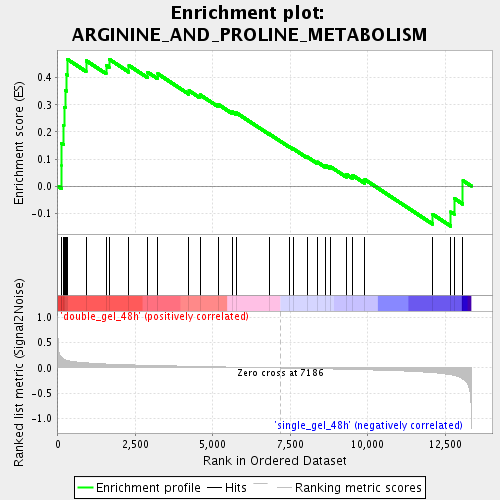
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | ARGININE_AND_PROLINE_METABOLISM |
| Enrichment Score (ES) | 0.4685453 |
| Normalized Enrichment Score (NES) | 1.53487 |
| Nominal p-value | 0.009803922 |
| FDR q-value | 0.12649292 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ABP1 | ABP1 Entrez, Source | amiloride binding protein 1 (amine oxidase (copper-containing)) | 113 | 0.213 | 0.0760 | Yes |
| 2 | ARG1 | ARG1 Entrez, Source | arginase, liver | 117 | 0.209 | 0.1585 | Yes |
| 3 | OTC | OTC Entrez, Source | ornithine carbamoyltransferase | 183 | 0.178 | 0.2240 | Yes |
| 4 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 197 | 0.173 | 0.2918 | Yes |
| 5 | CPS1 | CPS1 Entrez, Source | carbamoyl-phosphate synthetase 1, mitochondrial | 236 | 0.162 | 0.3533 | Yes |
| 6 | ASL | ASL Entrez, Source | argininosuccinate lyase | 273 | 0.154 | 0.4117 | Yes |
| 7 | AGMAT | AGMAT Entrez, Source | agmatine ureohydrolase (agmatinase) | 312 | 0.148 | 0.4676 | Yes |
| 8 | GAMT | GAMT Entrez, Source | guanidinoacetate N-methyltransferase | 910 | 0.102 | 0.4632 | Yes |
| 9 | AOC3 | AOC3 Entrez, Source | amine oxidase, copper containing 3 (vascular adhesion protein 1) | 1556 | 0.078 | 0.4458 | Yes |
| 10 | ALDH1A1 | ALDH1A1 Entrez, Source | aldehyde dehydrogenase 1 family, member A1 | 1653 | 0.076 | 0.4685 | Yes |
| 11 | NOS3 | NOS3 Entrez, Source | nitric oxide synthase 3 (endothelial cell) | 2292 | 0.062 | 0.4453 | No |
| 12 | ALDH1A2 | ALDH1A2 Entrez, Source | aldehyde dehydrogenase 1 family, member A2 | 2896 | 0.052 | 0.4205 | No |
| 13 | NOS1 | NOS1 Entrez, Source | nitric oxide synthase 1 (neuronal) | 3225 | 0.047 | 0.4145 | No |
| 14 | GATM | GATM Entrez, Source | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 4229 | 0.033 | 0.3523 | No |
| 15 | P4HB | P4HB Entrez, Source | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), beta polypeptide | 4586 | 0.029 | 0.3370 | No |
| 16 | MAOA | MAOA Entrez, Source | monoamine oxidase A | 5177 | 0.022 | 0.3013 | No |
| 17 | AMD1 | AMD1 Entrez, Source | adenosylmethionine decarboxylase 1 | 5628 | 0.017 | 0.2742 | No |
| 18 | CKM | CKM Entrez, Source | creatine kinase, muscle | 5748 | 0.015 | 0.2714 | No |
| 19 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 6812 | 0.004 | 0.1931 | No |
| 20 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 7461 | -0.003 | 0.1457 | No |
| 21 | ODC1 | ODC1 Entrez, Source | ornithine decarboxylase 1 | 7596 | -0.005 | 0.1375 | No |
| 22 | OAT | OAT Entrez, Source | ornithine aminotransferase (gyrate atrophy) | 8046 | -0.010 | 0.1078 | No |
| 23 | GLUD1 | GLUD1 Entrez, Source | glutamate dehydrogenase 1 | 8367 | -0.014 | 0.0892 | No |
| 24 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 8630 | -0.017 | 0.0764 | No |
| 25 | MAOB | MAOB Entrez, Source | monoamine oxidase B | 8794 | -0.019 | 0.0717 | No |
| 26 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 9309 | -0.026 | 0.0434 | No |
| 27 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 9511 | -0.029 | 0.0399 | No |
| 28 | CKB | CKB Entrez, Source | creatine kinase, brain | 9906 | -0.035 | 0.0241 | No |
| 29 | ARG2 | ARG2 Entrez, Source | arginase, type II | 12097 | -0.091 | -0.1042 | No |
| 30 | ALDH1B1 | ALDH1B1 Entrez, Source | aldehyde dehydrogenase 1 family, member B1 | 12682 | -0.133 | -0.0954 | No |
| 31 | P4HA1 | P4HA1 Entrez, Source | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide I | 12791 | -0.146 | -0.0455 | No |
| 32 | SMS | SMS Entrez, Source | spermine synthase | 13073 | -0.219 | 0.0202 | No |