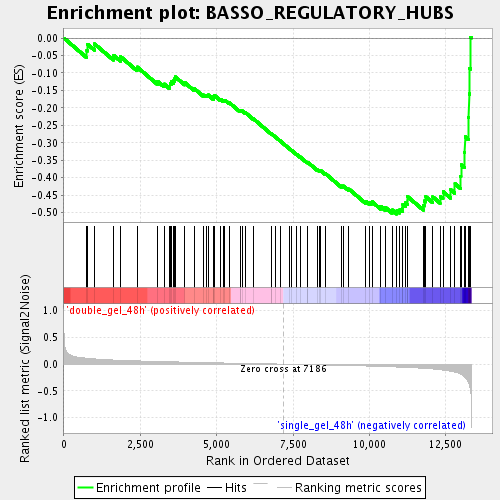
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | single_gel_48h |
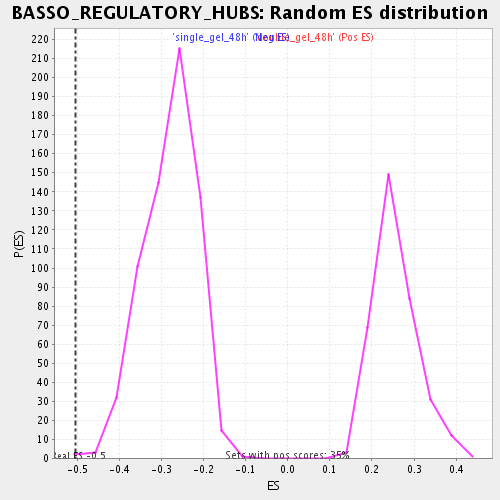
| GeneSet | BASSO_REGULATORY_HUBS |
| Enrichment Score (ES) | -0.50433564 |
| Normalized Enrichment Score (NES) | -1.8006498 |
| Nominal p-value | 0.0015360983 |
| FDR q-value | 0.04336547 |
| FWER p-Value | 0.975 |

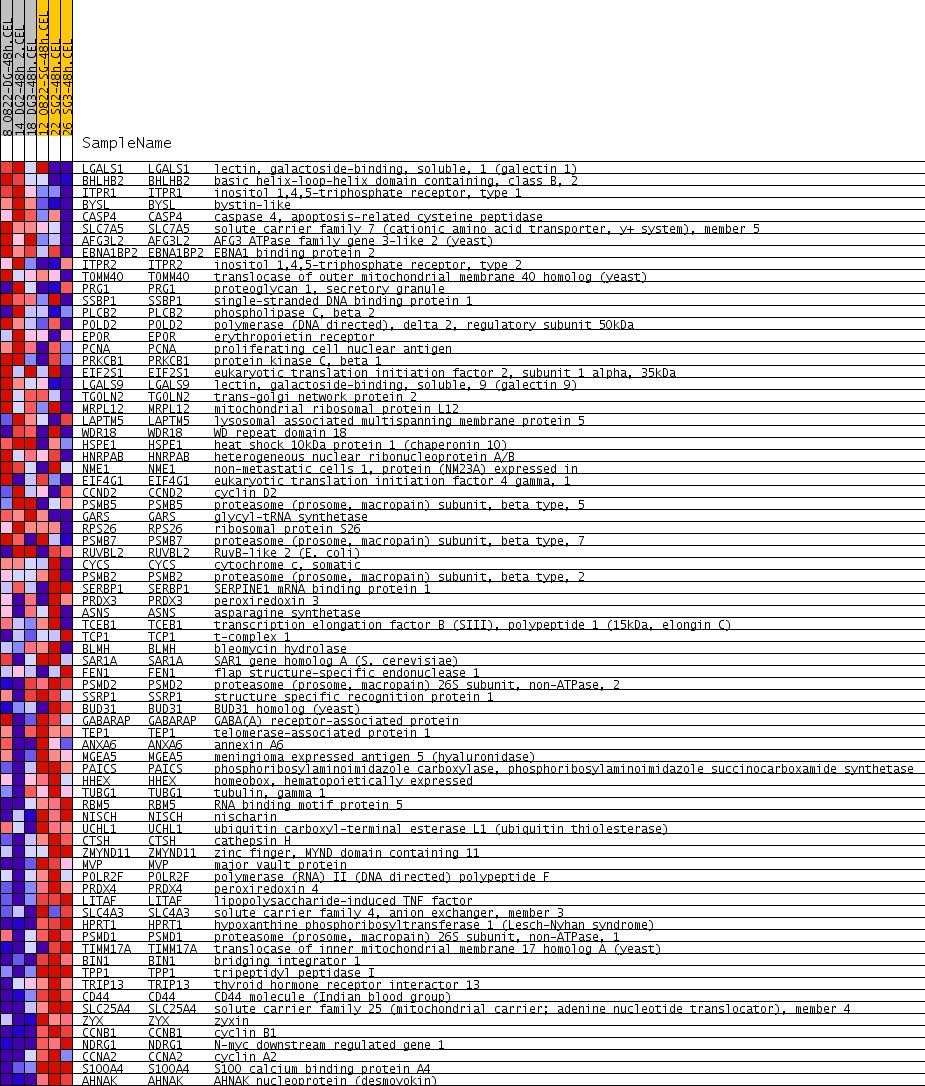
| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | LGALS1 | LGALS1 Entrez, Source | lectin, galactoside-binding, soluble, 1 (galectin 1) | 749 | 0.109 | -0.0361 | No |
| 2 | BHLHB2 | BHLHB2 Entrez, Source | basic helix-loop-helix domain containing, class B, 2 | 766 | 0.108 | -0.0172 | No |
| 3 | ITPR1 | ITPR1 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 1 | 1000 | 0.098 | -0.0165 | No |
| 4 | BYSL | BYSL Entrez, Source | bystin-like | 1620 | 0.077 | -0.0489 | No |
| 5 | CASP4 | CASP4 Entrez, Source | caspase 4, apoptosis-related cysteine peptidase | 1853 | 0.071 | -0.0532 | No |
| 6 | SLC7A5 | SLC7A5 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 2396 | 0.060 | -0.0828 | No |
| 7 | AFG3L2 | AFG3L2 Entrez, Source | AFG3 ATPase family gene 3-like 2 (yeast) | 3064 | 0.049 | -0.1239 | No |
| 8 | EBNA1BP2 | EBNA1BP2 Entrez, Source | EBNA1 binding protein 2 | 3279 | 0.046 | -0.1315 | No |
| 9 | ITPR2 | ITPR2 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 2 | 3468 | 0.043 | -0.1375 | No |
| 10 | TOMM40 | TOMM40 Entrez, Source | translocase of outer mitochondrial membrane 40 homolog (yeast) | 3471 | 0.043 | -0.1296 | No |
| 11 | PRG1 | PRG1 Entrez, Source | proteoglycan 1, secretory granule | 3518 | 0.043 | -0.1251 | No |
| 12 | SSBP1 | SSBP1 Entrez, Source | single-stranded DNA binding protein 1 | 3588 | 0.042 | -0.1225 | No |
| 13 | PLCB2 | PLCB2 Entrez, Source | phospholipase C, beta 2 | 3617 | 0.042 | -0.1169 | No |
| 14 | POLD2 | POLD2 Entrez, Source | polymerase (DNA directed), delta 2, regulatory subunit 50kDa | 3642 | 0.041 | -0.1110 | No |
| 15 | EPOR | EPOR Entrez, Source | erythropoietin receptor | 3955 | 0.037 | -0.1276 | No |
| 16 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 4258 | 0.033 | -0.1442 | No |
| 17 | PRKCB1 | PRKCB1 Entrez, Source | protein kinase C, beta 1 | 4579 | 0.029 | -0.1629 | No |
| 18 | EIF2S1 | EIF2S1 Entrez, Source | eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa | 4657 | 0.028 | -0.1635 | No |
| 19 | LGALS9 | LGALS9 Entrez, Source | lectin, galactoside-binding, soluble, 9 (galectin 9) | 4715 | 0.027 | -0.1627 | No |
| 20 | TGOLN2 | TGOLN2 Entrez, Source | trans-golgi network protein 2 | 4893 | 0.025 | -0.1713 | No |
| 21 | MRPL12 | MRPL12 Entrez, Source | mitochondrial ribosomal protein L12 | 4897 | 0.025 | -0.1669 | No |
| 22 | LAPTM5 | LAPTM5 Entrez, Source | lysosomal associated multispanning membrane protein 5 | 4925 | 0.025 | -0.1643 | No |
| 23 | WDR18 | WDR18 Entrez, Source | WD repeat domain 18 | 5126 | 0.022 | -0.1752 | No |
| 24 | HSPE1 | HSPE1 Entrez, Source | heat shock 10kDa protein 1 (chaperonin 10) | 5218 | 0.021 | -0.1781 | No |
| 25 | HNRPAB | HNRPAB Entrez, Source | heterogeneous nuclear ribonucleoprotein A/B | 5269 | 0.021 | -0.1780 | No |
| 26 | NME1 | NME1 Entrez, Source | non-metastatic cells 1, protein (NM23A) expressed in | 5403 | 0.019 | -0.1844 | No |
| 27 | EIF4G1 | EIF4G1 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 1 | 5763 | 0.015 | -0.2086 | No |
| 28 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 5771 | 0.015 | -0.2063 | No |
| 29 | PSMB5 | PSMB5 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 5 | 5837 | 0.015 | -0.2085 | No |
| 30 | GARS | GARS Entrez, Source | glycyl-tRNA synthetase | 5929 | 0.014 | -0.2129 | No |
| 31 | RPS26 | RPS26 Entrez, Source | ribosomal protein S26 | 6193 | 0.011 | -0.2306 | No |
| 32 | PSMB7 | PSMB7 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 7 | 6790 | 0.004 | -0.2748 | No |
| 33 | RUVBL2 | RUVBL2 Entrez, Source | RuvB-like 2 (E. coli) | 6799 | 0.004 | -0.2746 | No |
| 34 | CYCS | CYCS Entrez, Source | cytochrome c, somatic | 6937 | 0.003 | -0.2844 | No |
| 35 | PSMB2 | PSMB2 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 2 | 7074 | 0.001 | -0.2944 | No |
| 36 | SERBP1 | SERBP1 Entrez, Source | SERPINE1 mRNA binding protein 1 | 7386 | -0.002 | -0.3175 | No |
| 37 | PRDX3 | PRDX3 Entrez, Source | peroxiredoxin 3 | 7459 | -0.003 | -0.3223 | No |
| 38 | ASNS | ASNS Entrez, Source | asparagine synthetase | 7614 | -0.005 | -0.3330 | No |
| 39 | TCEB1 | TCEB1 Entrez, Source | transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) | 7731 | -0.006 | -0.3406 | No |
| 40 | TCP1 | TCP1 Entrez, Source | t-complex 1 | 7956 | -0.009 | -0.3558 | No |
| 41 | BLMH | BLMH Entrez, Source | bleomycin hydrolase | 7987 | -0.009 | -0.3563 | No |
| 42 | SAR1A | SAR1A Entrez, Source | SAR1 gene homolog A (S. cerevisiae) | 8302 | -0.013 | -0.3775 | No |
| 43 | FEN1 | FEN1 Entrez, Source | flap structure-specific endonuclease 1 | 8375 | -0.014 | -0.3804 | No |
| 44 | PSMD2 | PSMD2 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | 8388 | -0.014 | -0.3787 | No |
| 45 | SSRP1 | SSRP1 Entrez, Source | structure specific recognition protein 1 | 8553 | -0.016 | -0.3880 | No |
| 46 | BUD31 | BUD31 Entrez, Source | BUD31 homolog (yeast) | 9084 | -0.023 | -0.4237 | No |
| 47 | GABARAP | GABARAP Entrez, Source | GABA(A) receptor-associated protein | 9143 | -0.024 | -0.4237 | No |
| 48 | TEP1 | TEP1 Entrez, Source | telomerase-associated protein 1 | 9322 | -0.026 | -0.4322 | No |
| 49 | ANXA6 | ANXA6 Entrez, Source | annexin A6 | 9885 | -0.035 | -0.4681 | No |
| 50 | MGEA5 | MGEA5 Entrez, Source | meningioma expressed antigen 5 (hyaluronidase) | 10016 | -0.037 | -0.4710 | No |
| 51 | PAICS | PAICS Entrez, Source | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | 10082 | -0.038 | -0.4688 | No |
| 52 | HHEX | HHEX Entrez, Source | homeobox, hematopoietically expressed | 10368 | -0.043 | -0.4823 | No |
| 53 | TUBG1 | TUBG1 Entrez, Source | tubulin, gamma 1 | 10526 | -0.046 | -0.4857 | No |
| 54 | RBM5 | RBM5 Entrez, Source | RNA binding motif protein 5 | 10740 | -0.050 | -0.4924 | No |
| 55 | NISCH | NISCH Entrez, Source | nischarin | 10900 | -0.054 | -0.4943 | Yes |
| 56 | UCHL1 | UCHL1 Entrez, Source | ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) | 10996 | -0.056 | -0.4911 | Yes |
| 57 | CTSH | CTSH Entrez, Source | cathepsin H | 11083 | -0.058 | -0.4868 | Yes |
| 58 | ZMYND11 | ZMYND11 Entrez, Source | zinc finger, MYND domain containing 11 | 11091 | -0.058 | -0.4766 | Yes |
| 59 | MVP | MVP Entrez, Source | major vault protein | 11178 | -0.060 | -0.4719 | Yes |
| 60 | POLR2F | POLR2F Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide F | 11240 | -0.061 | -0.4651 | Yes |
| 61 | PRDX4 | PRDX4 Entrez, Source | peroxiredoxin 4 | 11241 | -0.061 | -0.4538 | Yes |
| 62 | LITAF | LITAF Entrez, Source | lipopolysaccharide-induced TNF factor | 11763 | -0.078 | -0.4786 | Yes |
| 63 | SLC4A3 | SLC4A3 Entrez, Source | solute carrier family 4, anion exchanger, member 3 | 11799 | -0.079 | -0.4665 | Yes |
| 64 | HPRT1 | HPRT1 Entrez, Source | hypoxanthine phosphoribosyltransferase 1 (Lesch-Nyhan syndrome) | 11843 | -0.081 | -0.4547 | Yes |
| 65 | PSMD1 | PSMD1 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | 12072 | -0.090 | -0.4551 | Yes |
| 66 | TIMM17A | TIMM17A Entrez, Source | translocase of inner mitochondrial membrane 17 homolog A (yeast) | 12312 | -0.104 | -0.4536 | Yes |
| 67 | BIN1 | BIN1 Entrez, Source | bridging integrator 1 | 12408 | -0.112 | -0.4400 | Yes |
| 68 | TPP1 | TPP1 Entrez, Source | tripeptidyl peptidase I | 12658 | -0.131 | -0.4344 | Yes |
| 69 | TRIP13 | TRIP13 Entrez, Source | thyroid hormone receptor interactor 13 | 12798 | -0.147 | -0.4174 | Yes |
| 70 | CD44 | CD44 Entrez, Source | CD44 molecule (Indian blood group) | 12977 | -0.184 | -0.3966 | Yes |
| 71 | SLC25A4 | SLC25A4 Entrez, Source | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 | 12996 | -0.187 | -0.3631 | Yes |
| 72 | ZYX | ZYX Entrez, Source | zyxin | 13115 | -0.238 | -0.3276 | Yes |
| 73 | CCNB1 | CCNB1 Entrez, Source | cyclin B1 | 13126 | -0.245 | -0.2827 | Yes |
| 74 | NDRG1 | NDRG1 Entrez, Source | N-myc downstream regulated gene 1 | 13242 | -0.345 | -0.2271 | Yes |
| 75 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 13257 | -0.367 | -0.1598 | Yes |
| 76 | S100A4 | S100A4 Entrez, Source | S100 calcium binding protein A4 | 13279 | -0.400 | -0.0869 | Yes |
| 77 | AHNAK | AHNAK Entrez, Source | AHNAK nucleoprotein (desmoyokin) | 13311 | -0.491 | 0.0023 | Yes |