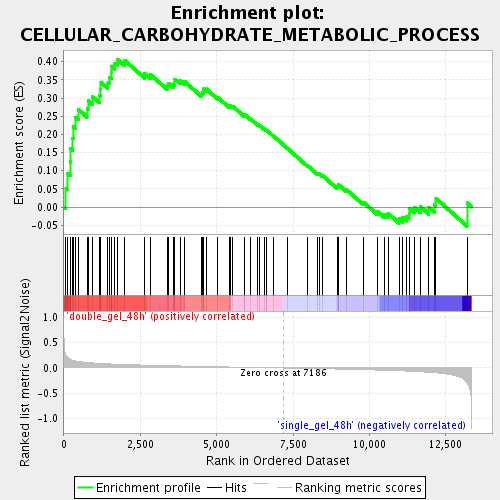
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | CELLULAR_CARBOHYDRATE_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.40688378 |
| Normalized Enrichment Score (NES) | 1.5670912 |
| Nominal p-value | 0.013586956 |
| FDR q-value | 0.112839796 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FBP1 | FBP1 Entrez, Source | fructose-1,6-bisphosphatase 1 | 67 | 0.255 | 0.0517 | Yes |
| 2 | ACLY | ACLY Entrez, Source | ATP citrate lyase | 127 | 0.202 | 0.0923 | Yes |
| 3 | PDK4 | PDK4 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 4 | 207 | 0.172 | 0.1245 | Yes |
| 4 | GCK | GCK Entrez, Source | glucokinase (hexokinase 4, maturity onset diabetes of the young 2) | 218 | 0.170 | 0.1614 | Yes |
| 5 | MIOX | MIOX Entrez, Source | myo-inositol oxygenase | 289 | 0.151 | 0.1897 | Yes |
| 6 | HPSE | HPSE Entrez, Source | heparanase | 311 | 0.148 | 0.2211 | Yes |
| 7 | GYS2 | GYS2 Entrez, Source | glycogen synthase 2 (liver) | 370 | 0.139 | 0.2476 | Yes |
| 8 | PFKFB1 | PFKFB1 Entrez, Source | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | 476 | 0.127 | 0.2679 | Yes |
| 9 | EPM2A | EPM2A Entrez, Source | epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) | 756 | 0.109 | 0.2710 | Yes |
| 10 | SLC25A10 | SLC25A10 Entrez, Source | solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 | 790 | 0.107 | 0.2924 | Yes |
| 11 | ALDOB | ALDOB Entrez, Source | aldolase B, fructose-bisphosphate | 941 | 0.100 | 0.3033 | Yes |
| 12 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 1159 | 0.091 | 0.3071 | Yes |
| 13 | NDST1 | NDST1 Entrez, Source | N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 | 1207 | 0.089 | 0.3235 | Yes |
| 14 | GAD2 | GAD2 Entrez, Source | glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) | 1212 | 0.089 | 0.3430 | Yes |
| 15 | GNE | GNE Entrez, Source | glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase | 1441 | 0.081 | 0.3439 | Yes |
| 16 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 1493 | 0.080 | 0.3578 | Yes |
| 17 | UGDH | UGDH Entrez, Source | UDP-glucose dehydrogenase | 1562 | 0.078 | 0.3700 | Yes |
| 18 | CHST3 | CHST3 Entrez, Source | carbohydrate (chondroitin 6) sulfotransferase 3 | 1564 | 0.078 | 0.3872 | Yes |
| 19 | NANP | NANP Entrez, Source | N-acetylneuraminic acid phosphatase | 1668 | 0.075 | 0.3962 | Yes |
| 20 | ACN9 | ACN9 Entrez, Source | ACN9 homolog (S. cerevisiae) | 1743 | 0.073 | 0.4069 | Yes |
| 21 | LALBA | LALBA Entrez, Source | lactalbumin, alpha- | 1989 | 0.068 | 0.4036 | No |
| 22 | LECT1 | LECT1 Entrez, Source | leukocyte cell derived chemotaxin 1 | 2642 | 0.056 | 0.3670 | No |
| 23 | GMDS | GMDS Entrez, Source | GDP-mannose 4,6-dehydratase | 2826 | 0.053 | 0.3650 | No |
| 24 | FPGT | FPGT Entrez, Source | fucose-1-phosphate guanylyltransferase | 3376 | 0.045 | 0.3336 | No |
| 25 | GFPT1 | GFPT1 Entrez, Source | glutamine-fructose-6-phosphate transaminase 1 | 3414 | 0.044 | 0.3406 | No |
| 26 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 3579 | 0.042 | 0.3375 | No |
| 27 | PYGM | PYGM Entrez, Source | phosphorylase, glycogen; muscle (McArdle syndrome, glycogen storage disease type V) | 3629 | 0.041 | 0.3431 | No |
| 28 | GBA2 | GBA2 Entrez, Source | glucosidase, beta (bile acid) 2 | 3630 | 0.041 | 0.3523 | No |
| 29 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 3799 | 0.039 | 0.3483 | No |
| 30 | IDH1 | IDH1 Entrez, Source | isocitrate dehydrogenase 1 (NADP+), soluble | 3953 | 0.037 | 0.3451 | No |
| 31 | GFPT2 | GFPT2 Entrez, Source | glutamine-fructose-6-phosphate transaminase 2 | 4497 | 0.030 | 0.3108 | No |
| 32 | FBP2 | FBP2 Entrez, Source | fructose-1,6-bisphosphatase 2 | 4526 | 0.030 | 0.3153 | No |
| 33 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 4559 | 0.029 | 0.3194 | No |
| 34 | PTEN | PTEN Entrez, Source | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 4572 | 0.029 | 0.3250 | No |
| 35 | ACO1 | ACO1 Entrez, Source | aconitase 1, soluble | 4655 | 0.028 | 0.3250 | No |
| 36 | XYLT1 | XYLT1 Entrez, Source | xylosyltransferase I | 5029 | 0.023 | 0.3022 | No |
| 37 | SDHD | SDHD Entrez, Source | succinate dehydrogenase complex, subunit D, integral membrane protein | 5418 | 0.019 | 0.2772 | No |
| 38 | GAD1 | GAD1 Entrez, Source | glutamate decarboxylase 1 (brain, 67kDa) | 5455 | 0.019 | 0.2786 | No |
| 39 | SDHC | SDHC Entrez, Source | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | 5522 | 0.018 | 0.2777 | No |
| 40 | HK1 | HK1 Entrez, Source | hexokinase 1 | 5904 | 0.014 | 0.2520 | No |
| 41 | TREH | TREH Entrez, Source | trehalase (brush-border membrane glycoprotein) | 5920 | 0.014 | 0.2539 | No |
| 42 | IDH3B | IDH3B Entrez, Source | isocitrate dehydrogenase 3 (NAD+) beta | 6092 | 0.012 | 0.2437 | No |
| 43 | CHST12 | CHST12 Entrez, Source | carbohydrate (chondroitin 4) sulfotransferase 12 | 6349 | 0.009 | 0.2264 | No |
| 44 | ALDOC | ALDOC Entrez, Source | aldolase C, fructose-bisphosphate | 6404 | 0.008 | 0.2243 | No |
| 45 | PPARGC1A | PPARGC1A Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, alpha | 6549 | 0.007 | 0.2150 | No |
| 46 | IPPK | IPPK Entrez, Source | inositol 1,3,4,5,6-pentakisphosphate 2-kinase | 6623 | 0.006 | 0.2109 | No |
| 47 | GAPDHS | GAPDHS Entrez, Source | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | 6630 | 0.006 | 0.2118 | No |
| 48 | AKR1A1 | AKR1A1 Entrez, Source | aldo-keto reductase family 1, member A1 (aldehyde reductase) | 6869 | 0.003 | 0.1946 | No |
| 49 | PDK2 | PDK2 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 2 | 7313 | -0.002 | 0.1616 | No |
| 50 | GALNT5 | GALNT5 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) | 7975 | -0.009 | 0.1138 | No |
| 51 | PDK1 | PDK1 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 1 | 8304 | -0.013 | 0.0920 | No |
| 52 | IL17F | IL17F Entrez, Source | interleukin 17F | 8354 | -0.014 | 0.0913 | No |
| 53 | B4GALT7 | B4GALT7 Entrez, Source | xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) | 8468 | -0.015 | 0.0862 | No |
| 54 | UGP2 | UGP2 Entrez, Source | UDP-glucose pyrophosphorylase 2 | 8944 | -0.021 | 0.0551 | No |
| 55 | SLC35A3 | SLC35A3 Entrez, Source | solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 | 8981 | -0.022 | 0.0572 | No |
| 56 | ARPP-19 | ARPP-19 Entrez, Source | - | 8985 | -0.022 | 0.0618 | No |
| 57 | ST3GAL6 | ST3GAL6 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 6 | 9231 | -0.025 | 0.0489 | No |
| 58 | GALK1 | GALK1 Entrez, Source | galactokinase 1 | 9799 | -0.033 | 0.0136 | No |
| 59 | CHST1 | CHST1 Entrez, Source | carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | 10266 | -0.041 | -0.0125 | No |
| 60 | GALT | GALT Entrez, Source | galactose-1-phosphate uridylyltransferase | 10491 | -0.045 | -0.0194 | No |
| 61 | PYGB | PYGB Entrez, Source | phosphorylase, glycogen; brain | 10620 | -0.047 | -0.0185 | No |
| 62 | EXTL2 | EXTL2 Entrez, Source | exostoses (multiple)-like 2 | 10968 | -0.055 | -0.0324 | No |
| 63 | CHIA | CHIA Entrez, Source | chitinase, acidic | 11088 | -0.058 | -0.0285 | No |
| 64 | GAA | GAA Entrez, Source | glucosidase, alpha; acid (Pompe disease, glycogen storage disease type II) | 11222 | -0.061 | -0.0250 | No |
| 65 | PFKM | PFKM Entrez, Source | phosphofructokinase, muscle | 11295 | -0.062 | -0.0166 | No |
| 66 | GNS | GNS Entrez, Source | glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID) | 11302 | -0.062 | -0.0032 | No |
| 67 | NAGK | NAGK Entrez, Source | N-acetylglucosamine kinase | 11479 | -0.068 | -0.0014 | No |
| 68 | GSK3B | GSK3B Entrez, Source | glycogen synthase kinase 3 beta | 11670 | -0.074 | 0.0008 | No |
| 69 | ALDOA | ALDOA Entrez, Source | aldolase A, fructose-bisphosphate | 11945 | -0.085 | -0.0010 | No |
| 70 | PFKL | PFKL Entrez, Source | phosphofructokinase, liver | 12126 | -0.093 | 0.0061 | No |
| 71 | GUSB | GUSB Entrez, Source | glucuronidase, beta | 12176 | -0.096 | 0.0236 | No |
| 72 | ST3GAL2 | ST3GAL2 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 2 | 13192 | -0.289 | 0.0113 | No |

