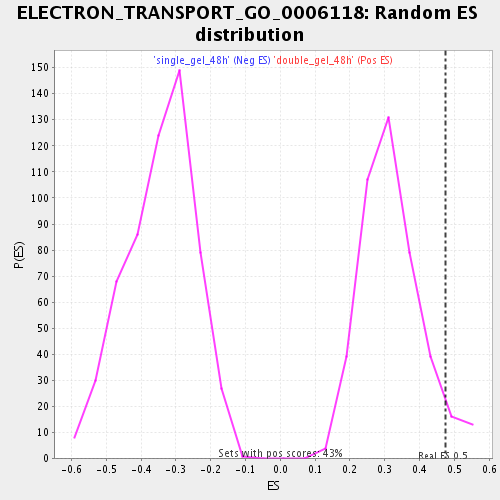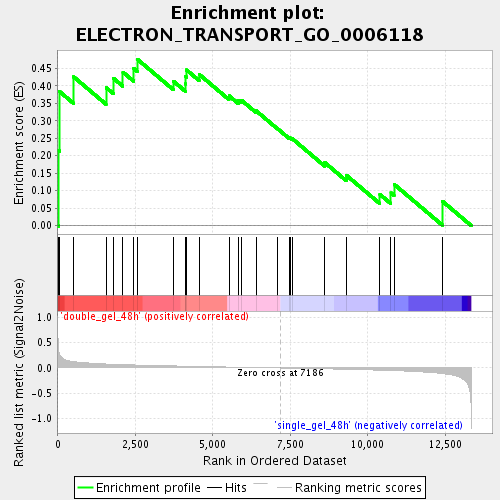
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | ELECTRON_TRANSPORT_GO_0006118 |
| Enrichment Score (ES) | 0.47609437 |
| Normalized Enrichment Score (NES) | 1.4912435 |
| Nominal p-value | 0.05140187 |
| FDR q-value | 0.15231523 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ADH7 | ADH7 Entrez, Source | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | 19 | 0.354 | 0.2153 | Yes |
| 2 | CDO1 | CDO1 Entrez, Source | cysteine dioxygenase, type I | 54 | 0.279 | 0.3835 | Yes |
| 3 | CYP1B1 | CYP1B1 Entrez, Source | cytochrome P450, family 1, subfamily B, polypeptide 1 | 506 | 0.124 | 0.4258 | Yes |
| 4 | AOC3 | AOC3 Entrez, Source | amine oxidase, copper containing 3 (vascular adhesion protein 1) | 1556 | 0.078 | 0.3948 | Yes |
| 5 | DHRS4 | DHRS4 Entrez, Source | dehydrogenase/reductase (SDR family) member 4 | 1791 | 0.072 | 0.4216 | Yes |
| 6 | AKR1D1 | AKR1D1 Entrez, Source | aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) | 2098 | 0.066 | 0.4390 | Yes |
| 7 | LOXL1 | LOXL1 Entrez, Source | lysyl oxidase-like 1 | 2447 | 0.060 | 0.4494 | Yes |
| 8 | POR | POR Entrez, Source | P450 (cytochrome) oxidoreductase | 2561 | 0.058 | 0.4761 | Yes |
| 9 | MTRR | MTRR Entrez, Source | 5-methyltetrahydrofolate-homocysteine methyltransferase reductase | 3727 | 0.040 | 0.4132 | No |
| 10 | ALDH5A1 | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 4114 | 0.035 | 0.4056 | No |
| 11 | PDIA5 | PDIA5 Entrez, Source | protein disulfide isomerase family A, member 5 | 4124 | 0.035 | 0.4262 | No |
| 12 | FDXR | FDXR Entrez, Source | ferredoxin reductase | 4149 | 0.034 | 0.4455 | No |
| 13 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 4559 | 0.029 | 0.4327 | No |
| 14 | SDHC | SDHC Entrez, Source | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | 5522 | 0.018 | 0.3715 | No |
| 15 | TXNL1 | TXNL1 Entrez, Source | thioredoxin-like 1 | 5826 | 0.015 | 0.3577 | No |
| 16 | GPX4 | GPX4 Entrez, Source | glutathione peroxidase 4 (phospholipid hydroperoxidase) | 5917 | 0.014 | 0.3593 | No |
| 17 | UQCRC1 | UQCRC1 Entrez, Source | ubiquinol-cytochrome c reductase core protein I | 6394 | 0.009 | 0.3288 | No |
| 18 | RETSAT | RETSAT Entrez, Source | retinol saturase (all-trans-retinol 13,14-reductase) | 7090 | 0.001 | 0.2772 | No |
| 19 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 7461 | -0.003 | 0.2514 | No |
| 20 | BLVRA | BLVRA Entrez, Source | biliverdin reductase A | 7498 | -0.004 | 0.2510 | No |
| 21 | NOX4 | NOX4 Entrez, Source | NADPH oxidase 4 | 7556 | -0.004 | 0.2494 | No |
| 22 | CYB5R2 | CYB5R2 Entrez, Source | cytochrome b5 reductase 2 | 8613 | -0.017 | 0.1806 | No |
| 23 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 9309 | -0.026 | 0.1443 | No |
| 24 | SURF1 | SURF1 Entrez, Source | surfeit 1 | 10391 | -0.043 | 0.0896 | No |
| 25 | SPR | SPR Entrez, Source | sepiapterin reductase (7,8-dihydrobiopterin:NADP+ oxidoreductase) | 10749 | -0.051 | 0.0938 | No |
| 26 | DUOX2 | DUOX2 Entrez, Source | dual oxidase 2 | 10851 | -0.053 | 0.1184 | No |
| 27 | PLOD1 | PLOD1 Entrez, Source | procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | 12413 | -0.112 | 0.0698 | No |