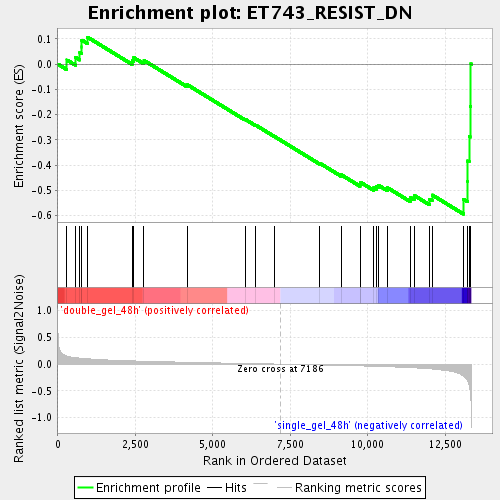
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | single_gel_48h |
| GeneSet | ET743_RESIST_DN |
| Enrichment Score (ES) | -0.5943755 |
| Normalized Enrichment Score (NES) | -1.7734376 |
| Nominal p-value | 0.0017452007 |
| FDR q-value | 0.047087125 |
| FWER p-Value | 0.992 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PPAP2B | PPAP2B Entrez, Source | phosphatidic acid phosphatase type 2B | 292 | 0.150 | 0.0165 | No |
| 2 | HSPA5 | HSPA5 Entrez, Source | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | 567 | 0.120 | 0.0267 | No |
| 3 | NOTCH3 | NOTCH3 Entrez, Source | Notch homolog 3 (Drosophila) | 706 | 0.111 | 0.0448 | No |
| 4 | BHLHB2 | BHLHB2 Entrez, Source | basic helix-loop-helix domain containing, class B, 2 | 766 | 0.108 | 0.0681 | No |
| 5 | CALR | CALR Entrez, Source | calreticulin | 768 | 0.108 | 0.0957 | No |
| 6 | DLC1 | DLC1 Entrez, Source | deleted in liver cancer 1 | 960 | 0.099 | 0.1067 | No |
| 7 | SLC7A5 | SLC7A5 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 2396 | 0.060 | 0.0143 | No |
| 8 | LOXL1 | LOXL1 Entrez, Source | lysyl oxidase-like 1 | 2447 | 0.060 | 0.0258 | No |
| 9 | SLC3A2 | SLC3A2 Entrez, Source | solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 | 2777 | 0.054 | 0.0148 | No |
| 10 | FSTL1 | FSTL1 Entrez, Source | follistatin-like 1 | 4179 | 0.034 | -0.0817 | No |
| 11 | NID1 | NID1 Entrez, Source | nidogen 1 | 6040 | 0.012 | -0.2183 | No |
| 12 | COL3A1 | COL3A1 Entrez, Source | collagen, type III, alpha 1 (Ehlers-Danlos syndrome type IV, autosomal dominant) | 6389 | 0.009 | -0.2422 | No |
| 13 | CSNK2A1 | CSNK2A1 Entrez, Source | casein kinase 2, alpha 1 polypeptide | 6997 | 0.002 | -0.2873 | No |
| 14 | SIRPA | SIRPA Entrez, Source | signal-regulatory protein alpha | 8454 | -0.015 | -0.3929 | No |
| 15 | GABRA5 | GABRA5 Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, alpha 5 | 9139 | -0.024 | -0.4382 | No |
| 16 | RND3 | RND3 Entrez, Source | Rho family GTPase 3 | 9751 | -0.033 | -0.4757 | No |
| 17 | PLXNB2 | PLXNB2 Entrez, Source | plexin B2 | 9766 | -0.033 | -0.4684 | No |
| 18 | GPI | GPI Entrez, Source | glucose phosphate isomerase | 10196 | -0.040 | -0.4904 | No |
| 19 | PKN1 | PKN1 Entrez, Source | protein kinase N1 | 10278 | -0.041 | -0.4860 | No |
| 20 | CNTN2 | CNTN2 Entrez, Source | contactin 2 (axonal) | 10343 | -0.042 | -0.4800 | No |
| 21 | CBLB | CBLB Entrez, Source | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | 10642 | -0.048 | -0.4902 | No |
| 22 | BCAM | BCAM Entrez, Source | basal cell adhesion molecule (Lutheran blood group) | 11372 | -0.064 | -0.5285 | No |
| 23 | PTTG1IP | PTTG1IP Entrez, Source | pituitary tumor-transforming 1 interacting protein | 11500 | -0.068 | -0.5206 | No |
| 24 | CDH13 | CDH13 Entrez, Source | cadherin 13, H-cadherin (heart) | 11996 | -0.087 | -0.5356 | No |
| 25 | ACTA2 | ACTA2 Entrez, Source | actin, alpha 2, smooth muscle, aorta | 12081 | -0.091 | -0.5187 | No |
| 26 | PRNP | PRNP Entrez, Source | prion protein (p27-30) (Creutzfeldt-Jakob disease, Gerstmann-Strausler-Scheinker syndrome, fatal familial insomnia) | 13090 | -0.227 | -0.5364 | Yes |
| 27 | SLC16A3 | SLC16A3 Entrez, Source | solute carrier family 16, member 3 (monocarboxylic acid transporter 4) | 13219 | -0.314 | -0.4657 | Yes |
| 28 | CITED2 | CITED2 Entrez, Source | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | 13226 | -0.322 | -0.3838 | Yes |
| 29 | ADM | ADM Entrez, Source | adrenomedullin | 13274 | -0.393 | -0.2867 | Yes |
| 30 | IGFBP7 | IGFBP7 Entrez, Source | insulin-like growth factor binding protein 7 | 13304 | -0.474 | -0.1676 | Yes |
| 31 | ANKRD1 | ANKRD1 Entrez, Source | ankyrin repeat domain 1 (cardiac muscle) | 13330 | -0.666 | 0.0009 | Yes |