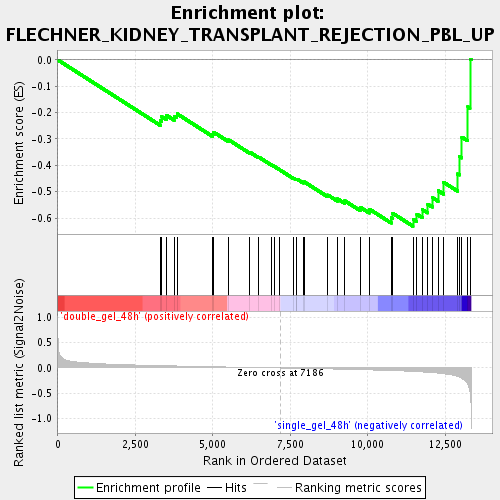
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | single_gel_48h |
| GeneSet | FLECHNER_KIDNEY_TRANSPLANT_REJECTION_PBL_UP |
| Enrichment Score (ES) | -0.6312581 |
| Normalized Enrichment Score (NES) | -1.9584297 |
| Nominal p-value | 0.0017301039 |
| FDR q-value | 0.01406981 |
| FWER p-Value | 0.373 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | OTUD4 | OTUD4 Entrez, Source | OTU domain containing 4 | 3307 | 0.046 | -0.2304 | No |
| 2 | TXNRD1 | TXNRD1 Entrez, Source | thioredoxin reductase 1 | 3342 | 0.045 | -0.2150 | No |
| 3 | HNRPA3 | HNRPA3 Entrez, Source | heterogeneous nuclear ribonucleoprotein A3 | 3502 | 0.043 | -0.2099 | No |
| 4 | CANX | CANX Entrez, Source | calnexin | 3753 | 0.040 | -0.2129 | No |
| 5 | SERPINE2 | SERPINE2 Entrez, Source | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 | 3843 | 0.039 | -0.2042 | No |
| 6 | PTPN12 | PTPN12 Entrez, Source | protein tyrosine phosphatase, non-receptor type 12 | 5000 | 0.024 | -0.2816 | No |
| 7 | CDKN1C | CDKN1C Entrez, Source | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 5020 | 0.024 | -0.2737 | No |
| 8 | ZFP161 | ZFP161 Entrez, Source | zinc finger protein 161 homolog (mouse) | 5513 | 0.018 | -0.3034 | No |
| 9 | ANP32E | ANP32E Entrez, Source | acidic (leucine-rich) nuclear phosphoprotein 32 family, member E | 6197 | 0.011 | -0.3504 | No |
| 10 | TSC22D3 | TSC22D3 Entrez, Source | TSC22 domain family, member 3 | 6484 | 0.008 | -0.3688 | No |
| 11 | ARF6 | ARF6 Entrez, Source | ADP-ribosylation factor 6 | 6899 | 0.003 | -0.3987 | No |
| 12 | TAX1BP1 | TAX1BP1 Entrez, Source | Tax1 (human T-cell leukemia virus type I) binding protein 1 | 6983 | 0.002 | -0.4041 | No |
| 13 | TMED10 | TMED10 Entrez, Source | transmembrane emp24-like trafficking protein 10 (yeast) | 7149 | 0.000 | -0.4164 | No |
| 14 | ICAM2 | ICAM2 Entrez, Source | intercellular adhesion molecule 2 | 7616 | -0.005 | -0.4494 | No |
| 15 | XRCC5 | XRCC5 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining; Ku autoantigen, 80kDa) | 7688 | -0.006 | -0.4525 | No |
| 16 | PSMD6 | PSMD6 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 | 7715 | -0.006 | -0.4521 | No |
| 17 | AGPAT1 | AGPAT1 Entrez, Source | 1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) | 7919 | -0.009 | -0.4640 | No |
| 18 | MTDH | MTDH Entrez, Source | metadherin | 7951 | -0.009 | -0.4627 | No |
| 19 | DAXX | DAXX Entrez, Source | death-associated protein 6 | 8710 | -0.018 | -0.5125 | No |
| 20 | CDK7 | CDK7 Entrez, Source | cyclin-dependent kinase 7 (MO15 homolog, Xenopus laevis, cdk-activating kinase) | 9024 | -0.022 | -0.5273 | No |
| 21 | B2M | B2M Entrez, Source | beta-2-microglobulin | 9260 | -0.025 | -0.5348 | No |
| 22 | KNS2 | KNS2 Entrez, Source | kinesin 2 | 9759 | -0.033 | -0.5592 | No |
| 23 | NFYA | NFYA Entrez, Source | nuclear transcription factor Y, alpha | 10052 | -0.038 | -0.5662 | No |
| 24 | PARP1 | PARP1 Entrez, Source | poly (ADP-ribose) polymerase family, member 1 | 10758 | -0.051 | -0.5991 | No |
| 25 | ARHGEF1 | ARHGEF1 Entrez, Source | Rho guanine nucleotide exchange factor (GEF) 1 | 10799 | -0.052 | -0.5816 | No |
| 26 | SPINT2 | SPINT2 Entrez, Source | serine peptidase inhibitor, Kunitz type, 2 | 11461 | -0.067 | -0.6047 | Yes |
| 27 | SHOC2 | SHOC2 Entrez, Source | soc-2 suppressor of clear homolog (C. elegans) | 11584 | -0.071 | -0.5856 | Yes |
| 28 | LITAF | LITAF Entrez, Source | lipopolysaccharide-induced TNF factor | 11763 | -0.078 | -0.5681 | Yes |
| 29 | DNPEP | DNPEP Entrez, Source | aspartyl aminopeptidase | 11936 | -0.084 | -0.5475 | Yes |
| 30 | LYN | LYN Entrez, Source | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | 12083 | -0.091 | -0.5224 | Yes |
| 31 | RPA2 | RPA2 Entrez, Source | replication protein A2, 32kDa | 12271 | -0.101 | -0.4963 | Yes |
| 32 | ANXA4 | ANXA4 Entrez, Source | annexin A4 | 12446 | -0.114 | -0.4642 | Yes |
| 33 | TAX1BP3 | TAX1BP3 Entrez, Source | Tax1 (human T-cell leukemia virus type I) binding protein 3 | 12900 | -0.165 | -0.4327 | Yes |
| 34 | CD47 | CD47 Entrez, Source | CD47 molecule | 12956 | -0.177 | -0.3664 | Yes |
| 35 | CCDC131 | CCDC131 Entrez, Source | coiled-coil domain containing 131 | 13035 | -0.200 | -0.2928 | Yes |
| 36 | PDLIM5 | PDLIM5 Entrez, Source | PDZ and LIM domain 5 | 13230 | -0.326 | -0.1781 | Yes |
| 37 | MCM6 | MCM6 Entrez, Source | MCM6 minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) | 13301 | -0.469 | 0.0031 | Yes |