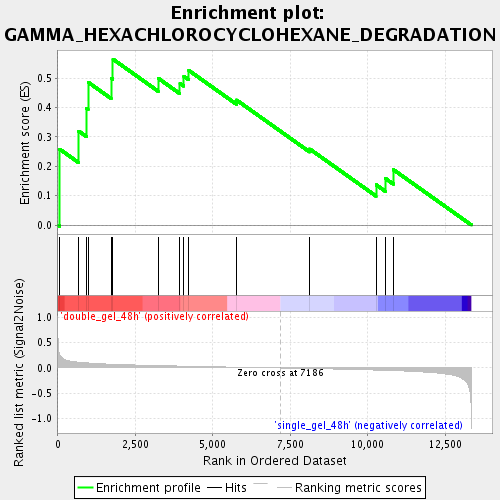
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | GAMMA_HEXACHLOROCYCLOHEXANE_DEGRADATION |
| Enrichment Score (ES) | 0.56517446 |
| Normalized Enrichment Score (NES) | 1.5231628 |
| Nominal p-value | 0.042986427 |
| FDR q-value | 0.12959054 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CYP51A1 | CYP51A1 Entrez, Source | cytochrome P450, family 51, subfamily A, polypeptide 1 | 56 | 0.277 | 0.2585 | Yes |
| 2 | CYP19A1 | CYP19A1 Entrez, Source | cytochrome P450, family 19, subfamily A, polypeptide 1 | 670 | 0.114 | 0.3202 | Yes |
| 3 | PON1 | PON1 Entrez, Source | paraoxonase 1 | 922 | 0.101 | 0.3971 | Yes |
| 4 | CYP3A4 | CYP3A4 Entrez, Source | cytochrome P450, family 3, subfamily A, polypeptide 4 | 975 | 0.099 | 0.4866 | Yes |
| 5 | CYP1A1 | CYP1A1 Entrez, Source | cytochrome P450, family 1, subfamily A, polypeptide 1 | 1719 | 0.074 | 0.5006 | Yes |
| 6 | CYP2C9 | CYP2C9 Entrez, Source | cytochrome P450, family 2, subfamily C, polypeptide 9 | 1777 | 0.073 | 0.5652 | Yes |
| 7 | CYP4B1 | CYP4B1 Entrez, Source | cytochrome P450, family 4, subfamily B, polypeptide 1 | 3238 | 0.047 | 0.4999 | No |
| 8 | CYP1A2 | CYP1A2 Entrez, Source | cytochrome P450, family 1, subfamily A, polypeptide 2 | 3939 | 0.037 | 0.4827 | No |
| 9 | ACP1 | ACP1 Entrez, Source | acid phosphatase 1, soluble | 4064 | 0.035 | 0.5070 | No |
| 10 | ALPL | ALPL Entrez, Source | alkaline phosphatase, liver/bone/kidney | 4222 | 0.034 | 0.5270 | No |
| 11 | ACP2 | ACP2 Entrez, Source | acid phosphatase 2, lysosomal | 5762 | 0.015 | 0.4261 | No |
| 12 | ALPI | ALPI Entrez, Source | alkaline phosphatase, intestinal | 8126 | -0.011 | 0.2591 | No |
| 13 | CYP2E1 | CYP2E1 Entrez, Source | cytochrome P450, family 2, subfamily E, polypeptide 1 | 10268 | -0.041 | 0.1372 | No |
| 14 | ACP5 | ACP5 Entrez, Source | acid phosphatase 5, tartrate resistant | 10569 | -0.046 | 0.1586 | No |
| 15 | ACPP | ACPP Entrez, Source | acid phosphatase, prostate | 10827 | -0.052 | 0.1887 | No |

