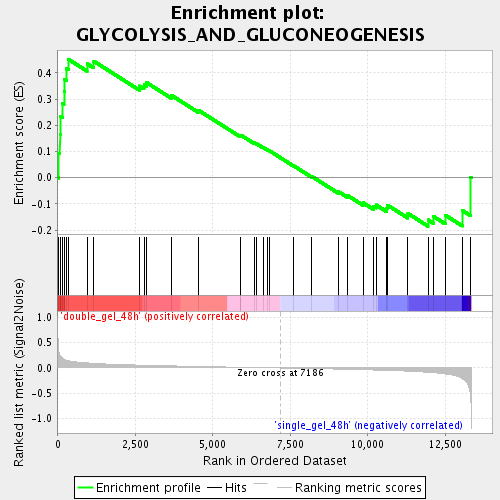
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | GLYCOLYSIS_AND_GLUCONEOGENESIS |
| Enrichment Score (ES) | 0.45190153 |
| Normalized Enrichment Score (NES) | 1.5376166 |
| Nominal p-value | 0.019323671 |
| FDR q-value | 0.12579893 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | G6PC | G6PC Entrez, Source | glucose-6-phosphatase, catalytic subunit | 26 | 0.337 | 0.0940 | Yes |
| 2 | FBP1 | FBP1 Entrez, Source | fructose-1,6-bisphosphatase 1 | 67 | 0.255 | 0.1636 | Yes |
| 3 | PKLR | PKLR Entrez, Source | pyruvate kinase, liver and RBC | 74 | 0.247 | 0.2334 | Yes |
| 4 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 148 | 0.193 | 0.2828 | Yes |
| 5 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 197 | 0.173 | 0.3285 | Yes |
| 6 | GCK | GCK Entrez, Source | glucokinase (hexokinase 4, maturity onset diabetes of the young 2) | 218 | 0.170 | 0.3753 | Yes |
| 7 | PC | PC Entrez, Source | pyruvate carboxylase | 270 | 0.155 | 0.4154 | Yes |
| 8 | PDHA2 | PDHA2 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 2 | 332 | 0.144 | 0.4519 | Yes |
| 9 | ALDOB | ALDOB Entrez, Source | aldolase B, fructose-bisphosphate | 941 | 0.100 | 0.4346 | No |
| 10 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 1159 | 0.091 | 0.4441 | No |
| 11 | LDHAL6B | LDHAL6B Entrez, Source | lactate dehydrogenase A-like 6B | 2630 | 0.056 | 0.3497 | No |
| 12 | ENO2 | ENO2 Entrez, Source | enolase 2 (gamma, neuronal) | 2779 | 0.054 | 0.3539 | No |
| 13 | MDH1 | MDH1 Entrez, Source | malate dehydrogenase 1, NAD (soluble) | 2853 | 0.052 | 0.3633 | No |
| 14 | MDH2 | MDH2 Entrez, Source | malate dehydrogenase 2, NAD (mitochondrial) | 3660 | 0.041 | 0.3143 | No |
| 15 | FBP2 | FBP2 Entrez, Source | fructose-1,6-bisphosphatase 2 | 4526 | 0.030 | 0.2578 | No |
| 16 | DLD | DLD Entrez, Source | dihydrolipoamide dehydrogenase | 5876 | 0.014 | 0.1605 | No |
| 17 | HK1 | HK1 Entrez, Source | hexokinase 1 | 5904 | 0.014 | 0.1624 | No |
| 18 | ENO3 | ENO3 Entrez, Source | enolase 3 (beta, muscle) | 6328 | 0.009 | 0.1332 | No |
| 19 | ALDOC | ALDOC Entrez, Source | aldolase C, fructose-bisphosphate | 6404 | 0.008 | 0.1300 | No |
| 20 | GAPDHS | GAPDHS Entrez, Source | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | 6630 | 0.006 | 0.1148 | No |
| 21 | PDHA1 | PDHA1 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 1 | 6772 | 0.004 | 0.1055 | No |
| 22 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 6812 | 0.004 | 0.1037 | No |
| 23 | GAPDH | GAPDH Entrez, Source | glyceraldehyde-3-phosphate dehydrogenase | 7606 | -0.005 | 0.0455 | No |
| 24 | LDHA | LDHA Entrez, Source | lactate dehydrogenase A | 8185 | -0.012 | 0.0054 | No |
| 25 | PGAM2 | PGAM2 Entrez, Source | phosphoglycerate mutase 2 (muscle) | 9063 | -0.023 | -0.0541 | No |
| 26 | PGK1 | PGK1 Entrez, Source | phosphoglycerate kinase 1 | 9333 | -0.026 | -0.0668 | No |
| 27 | PGAM1 | PGAM1 Entrez, Source | phosphoglycerate mutase 1 (brain) | 9850 | -0.034 | -0.0959 | No |
| 28 | GPI | GPI Entrez, Source | glucose phosphate isomerase | 10196 | -0.040 | -0.1105 | No |
| 29 | TPI1 | TPI1 Entrez, Source | triosephosphate isomerase 1 | 10267 | -0.041 | -0.1041 | No |
| 30 | LDHC | LDHC Entrez, Source | lactate dehydrogenase C | 10601 | -0.047 | -0.1158 | No |
| 31 | HK2 | HK2 Entrez, Source | hexokinase 2 | 10644 | -0.048 | -0.1053 | No |
| 32 | PFKM | PFKM Entrez, Source | phosphofructokinase, muscle | 11295 | -0.062 | -0.1365 | No |
| 33 | ALDOA | ALDOA Entrez, Source | aldolase A, fructose-bisphosphate | 11945 | -0.085 | -0.1611 | No |
| 34 | PFKL | PFKL Entrez, Source | phosphofructokinase, liver | 12126 | -0.093 | -0.1483 | No |
| 35 | DLAT | DLAT Entrez, Source | dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) | 12499 | -0.117 | -0.1429 | No |
| 36 | LDHB | LDHB Entrez, Source | lactate dehydrogenase B | 13053 | -0.209 | -0.1251 | No |
| 37 | PFKP | PFKP Entrez, Source | phosphofructokinase, platelet | 13318 | -0.516 | 0.0018 | No |

