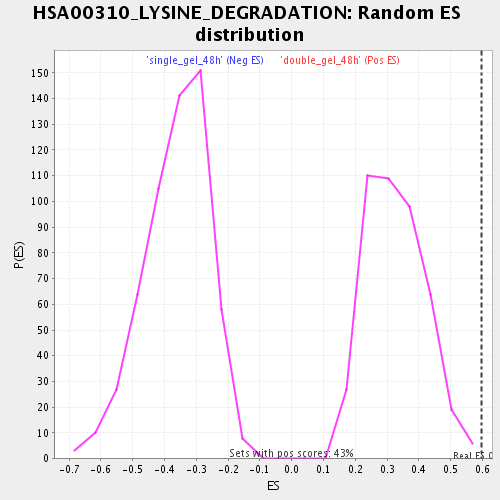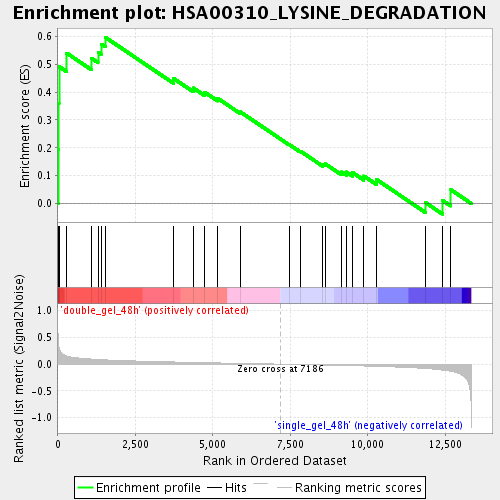
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | HSA00310_LYSINE_DEGRADATION |
| Enrichment Score (ES) | 0.5965609 |
| Normalized Enrichment Score (NES) | 1.8197008 |
| Nominal p-value | 0.0023094688 |
| FDR q-value | 0.028094502 |
| FWER p-Value | 0.85 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 6 | 0.447 | 0.1956 | Yes |
| 2 | ACAT2 | ACAT2 Entrez, Source | acetyl-Coenzyme A acetyltransferase 2 (acetoacetyl Coenzyme A thiolase) | 14 | 0.377 | 0.3604 | Yes |
| 3 | BBOX1 | BBOX1 Entrez, Source | butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 | 37 | 0.303 | 0.4916 | Yes |
| 4 | AADAT | AADAT Entrez, Source | aminoadipate aminotransferase | 283 | 0.152 | 0.5398 | Yes |
| 5 | AKR1B10 | AKR1B10 Entrez, Source | aldo-keto reductase family 1, member B10 (aldose reductase) | 1081 | 0.094 | 0.5213 | Yes |
| 6 | ACAT1 | ACAT1 Entrez, Source | acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | 1295 | 0.086 | 0.5431 | Yes |
| 7 | RDH11 | RDH11 Entrez, Source | retinol dehydrogenase 11 (all-trans/9-cis/11-cis) | 1395 | 0.083 | 0.5720 | Yes |
| 8 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 1528 | 0.079 | 0.5966 | Yes |
| 9 | SHMT2 | SHMT2 Entrez, Source | serine hydroxymethyltransferase 2 (mitochondrial) | 3728 | 0.040 | 0.4490 | No |
| 10 | HSD17B4 | HSD17B4 Entrez, Source | hydroxysteroid (17-beta) dehydrogenase 4 | 4370 | 0.032 | 0.4148 | No |
| 11 | SHMT1 | SHMT1 Entrez, Source | serine hydroxymethyltransferase 1 (soluble) | 4738 | 0.027 | 0.3992 | No |
| 12 | HADHA | HADHA Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 5156 | 0.022 | 0.3775 | No |
| 13 | DLST | DLST Entrez, Source | dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) | 5882 | 0.014 | 0.3292 | No |
| 14 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 7461 | -0.003 | 0.2121 | No |
| 15 | HADH | HADH Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase | 7824 | -0.007 | 0.1882 | No |
| 16 | HSD3B7 | HSD3B7 Entrez, Source | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 | 8545 | -0.016 | 0.1411 | No |
| 17 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 8630 | -0.017 | 0.1424 | No |
| 18 | PLOD3 | PLOD3 Entrez, Source | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 | 9156 | -0.024 | 0.1134 | No |
| 19 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 9309 | -0.026 | 0.1134 | No |
| 20 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 9511 | -0.029 | 0.1111 | No |
| 21 | TMLHE | TMLHE Entrez, Source | trimethyllysine hydroxylase, epsilon | 9874 | -0.034 | 0.0989 | No |
| 22 | ALDH7A1 | ALDH7A1 Entrez, Source | aldehyde dehydrogenase 7 family, member A1 | 10287 | -0.041 | 0.0860 | No |
| 23 | EHMT2 | EHMT2 Entrez, Source | euchromatic histone-lysine N-methyltransferase 2 | 11859 | -0.081 | 0.0038 | No |
| 24 | PLOD1 | PLOD1 Entrez, Source | procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | 12413 | -0.112 | 0.0114 | No |
| 25 | ALDH1B1 | ALDH1B1 Entrez, Source | aldehyde dehydrogenase 1 family, member B1 | 12682 | -0.133 | 0.0496 | No |