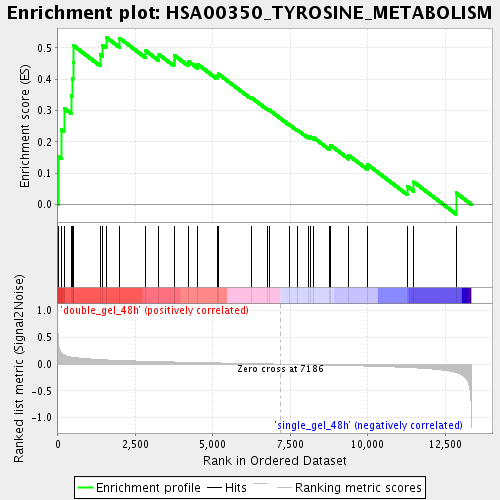
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
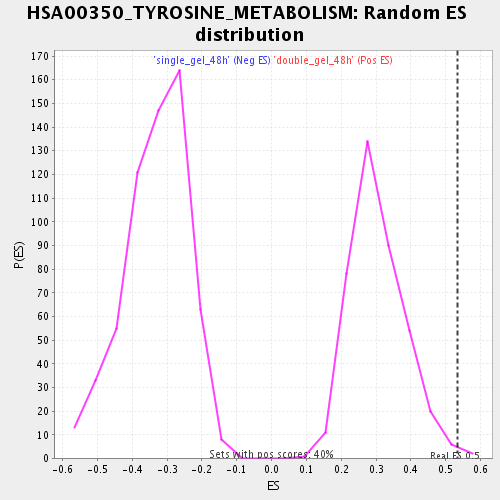
| GeneSet | HSA00350_TYROSINE_METABOLISM |
| Enrichment Score (ES) | 0.5338169 |
| Normalized Enrichment Score (NES) | 1.7516469 |
| Nominal p-value | 0.007575758 |
| FDR q-value | 0.0435968 |
| FWER p-Value | 0.972 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ADH7 | ADH7 Entrez, Source | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | 19 | 0.354 | 0.1525 | Yes |
| 2 | ABP1 | ABP1 Entrez, Source | amiloride binding protein 1 (amine oxidase (copper-containing)) | 113 | 0.213 | 0.2381 | Yes |
| 3 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 197 | 0.173 | 0.3073 | Yes |
| 4 | TAT | TAT Entrez, Source | tyrosine aminotransferase | 428 | 0.132 | 0.3475 | Yes |
| 5 | HGD | HGD Entrez, Source | homogentisate 1,2-dioxygenase (homogentisate oxidase) | 457 | 0.129 | 0.4015 | Yes |
| 6 | FAH | FAH Entrez, Source | fumarylacetoacetate hydrolase (fumarylacetoacetase) | 489 | 0.126 | 0.4539 | Yes |
| 7 | ADHFE1 | ADHFE1 Entrez, Source | alcohol dehydrogenase, iron containing, 1 | 507 | 0.124 | 0.5068 | Yes |
| 8 | AOX1 | AOX1 Entrez, Source | aldehyde oxidase 1 | 1365 | 0.084 | 0.4787 | Yes |
| 9 | ADH6 | ADH6 Entrez, Source | alcohol dehydrogenase 6 (class V) | 1443 | 0.081 | 0.5083 | Yes |
| 10 | AOC3 | AOC3 Entrez, Source | amine oxidase, copper containing 3 (vascular adhesion protein 1) | 1556 | 0.078 | 0.5338 | Yes |
| 11 | ADH1A | ADH1A Entrez, Source | alcohol dehydrogenase 1A (class I), alpha polypeptide | 1987 | 0.068 | 0.5313 | No |
| 12 | COMT | COMT Entrez, Source | catechol-O-methyltransferase | 2832 | 0.053 | 0.4908 | No |
| 13 | PNMT | PNMT Entrez, Source | phenylethanolamine N-methyltransferase | 3260 | 0.046 | 0.4788 | No |
| 14 | MYST3 | MYST3 Entrez, Source | MYST histone acetyltransferase (monocytic leukemia) 3 | 3748 | 0.040 | 0.4596 | No |
| 15 | TPO | TPO Entrez, Source | thyroid peroxidase | 3752 | 0.040 | 0.4767 | No |
| 16 | PRMT3 | PRMT3 Entrez, Source | protein arginine methyltransferase 3 | 4207 | 0.034 | 0.4572 | No |
| 17 | DDC | DDC Entrez, Source | dopa decarboxylase (aromatic L-amino acid decarboxylase) | 4512 | 0.030 | 0.4474 | No |
| 18 | CARM1 | CARM1 Entrez, Source | coactivator-associated arginine methyltransferase 1 | 5136 | 0.022 | 0.4102 | No |
| 19 | MAOA | MAOA Entrez, Source | monoamine oxidase A | 5177 | 0.022 | 0.4167 | No |
| 20 | ADH4 | ADH4 Entrez, Source | alcohol dehydrogenase 4 (class II), pi polypeptide | 6235 | 0.011 | 0.3418 | No |
| 21 | TH | TH Entrez, Source | tyrosine hydroxylase | 6759 | 0.005 | 0.3045 | No |
| 22 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 6812 | 0.004 | 0.3024 | No |
| 23 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 7461 | -0.003 | 0.2551 | No |
| 24 | PRMT2 | PRMT2 Entrez, Source | protein arginine methyltransferase 2 | 7736 | -0.006 | 0.2372 | No |
| 25 | LCMT1 | LCMT1 Entrez, Source | leucine carboxyl methyltransferase 1 | 8072 | -0.010 | 0.2165 | No |
| 26 | ECH1 | ECH1 Entrez, Source | enoyl Coenzyme A hydratase 1, peroxisomal | 8150 | -0.011 | 0.2156 | No |
| 27 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 8248 | -0.012 | 0.2137 | No |
| 28 | LCMT2 | LCMT2 Entrez, Source | leucine carboxyl methyltransferase 2 | 8774 | -0.019 | 0.1825 | No |
| 29 | MAOB | MAOB Entrez, Source | monoamine oxidase B | 8794 | -0.019 | 0.1894 | No |
| 30 | METTL6 | METTL6 Entrez, Source | methyltransferase like 6 | 9390 | -0.027 | 0.1566 | No |
| 31 | SH3GLB1 | SH3GLB1 Entrez, Source | SH3-domain GRB2-like endophilin B1 | 9998 | -0.037 | 0.1269 | No |
| 32 | MIF | MIF Entrez, Source | macrophage migration inhibitory factor (glycosylation-inhibiting factor) | 11278 | -0.062 | 0.0576 | No |
| 33 | DBH | DBH Entrez, Source | dopamine beta-hydroxylase (dopamine beta-monooxygenase) | 11484 | -0.068 | 0.0717 | No |
| 34 | ALDH3B1 | ALDH3B1 Entrez, Source | aldehyde dehydrogenase 3 family, member B1 | 12850 | -0.156 | 0.0370 | No |