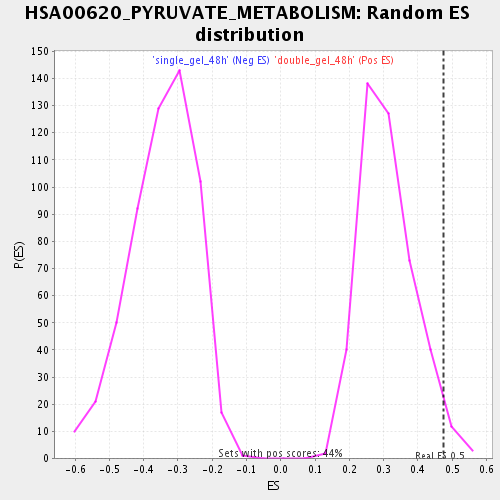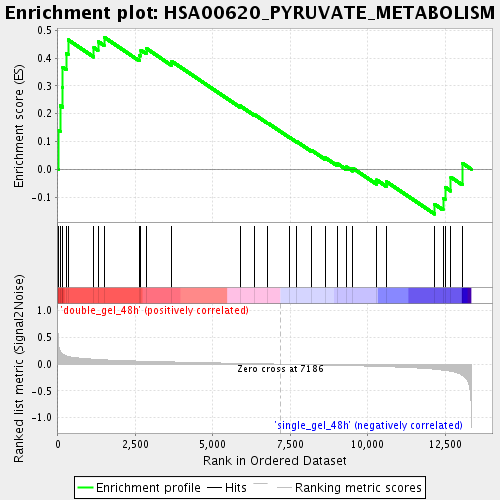
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | HSA00620_PYRUVATE_METABOLISM |
| Enrichment Score (ES) | 0.4746675 |
| Normalized Enrichment Score (NES) | 1.528987 |
| Nominal p-value | 0.025287356 |
| FDR q-value | 0.12757829 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ACAT2 | ACAT2 Entrez, Source | acetyl-Coenzyme A acetyltransferase 2 (acetoacetyl Coenzyme A thiolase) | 14 | 0.377 | 0.1404 | Yes |
| 2 | PKLR | PKLR Entrez, Source | pyruvate kinase, liver and RBC | 74 | 0.247 | 0.2286 | Yes |
| 3 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 148 | 0.193 | 0.2955 | Yes |
| 4 | ACACA | ACACA Entrez, Source | acetyl-Coenzyme A carboxylase alpha | 157 | 0.190 | 0.3662 | Yes |
| 5 | PC | PC Entrez, Source | pyruvate carboxylase | 270 | 0.155 | 0.4158 | Yes |
| 6 | PDHA2 | PDHA2 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 2 | 332 | 0.144 | 0.4654 | Yes |
| 7 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 1159 | 0.091 | 0.4375 | Yes |
| 8 | ACAT1 | ACAT1 Entrez, Source | acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | 1295 | 0.086 | 0.4596 | Yes |
| 9 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 1493 | 0.080 | 0.4747 | Yes |
| 10 | LDHAL6B | LDHAL6B Entrez, Source | lactate dehydrogenase A-like 6B | 2630 | 0.056 | 0.4105 | No |
| 11 | HAGH | HAGH Entrez, Source | hydroxyacylglutathione hydrolase | 2676 | 0.056 | 0.4280 | No |
| 12 | MDH1 | MDH1 Entrez, Source | malate dehydrogenase 1, NAD (soluble) | 2853 | 0.052 | 0.4345 | No |
| 13 | MDH2 | MDH2 Entrez, Source | malate dehydrogenase 2, NAD (mitochondrial) | 3660 | 0.041 | 0.3893 | No |
| 14 | DLD | DLD Entrez, Source | dihydrolipoamide dehydrogenase | 5876 | 0.014 | 0.2282 | No |
| 15 | ACACB | ACACB Entrez, Source | acetyl-Coenzyme A carboxylase beta | 6337 | 0.009 | 0.1971 | No |
| 16 | PDHA1 | PDHA1 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 1 | 6772 | 0.004 | 0.1662 | No |
| 17 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 7461 | -0.003 | 0.1157 | No |
| 18 | ACOT12 | ACOT12 Entrez, Source | acyl-CoA thioesterase 12 | 7714 | -0.006 | 0.0990 | No |
| 19 | LDHA | LDHA Entrez, Source | lactate dehydrogenase A | 8185 | -0.012 | 0.0681 | No |
| 20 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 8630 | -0.017 | 0.0412 | No |
| 21 | GLO1 | GLO1 Entrez, Source | glyoxalase I | 9022 | -0.022 | 0.0201 | No |
| 22 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 9309 | -0.026 | 0.0084 | No |
| 23 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 9511 | -0.029 | 0.0042 | No |
| 24 | ALDH7A1 | ALDH7A1 Entrez, Source | aldehyde dehydrogenase 7 family, member A1 | 10287 | -0.041 | -0.0386 | No |
| 25 | LDHC | LDHC Entrez, Source | lactate dehydrogenase C | 10601 | -0.047 | -0.0445 | No |
| 26 | LDHD | LDHD Entrez, Source | lactate dehydrogenase D | 12158 | -0.094 | -0.1260 | No |
| 27 | HAGHL | HAGHL Entrez, Source | hydroxyacylglutathione hydrolase-like | 12432 | -0.113 | -0.1040 | No |
| 28 | DLAT | DLAT Entrez, Source | dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) | 12499 | -0.117 | -0.0649 | No |
| 29 | ALDH1B1 | ALDH1B1 Entrez, Source | aldehyde dehydrogenase 1 family, member B1 | 12682 | -0.133 | -0.0287 | No |
| 30 | LDHB | LDHB Entrez, Source | lactate dehydrogenase B | 13053 | -0.209 | 0.0217 | No |