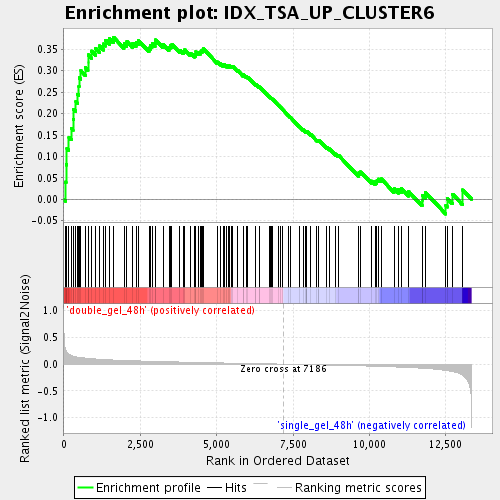
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
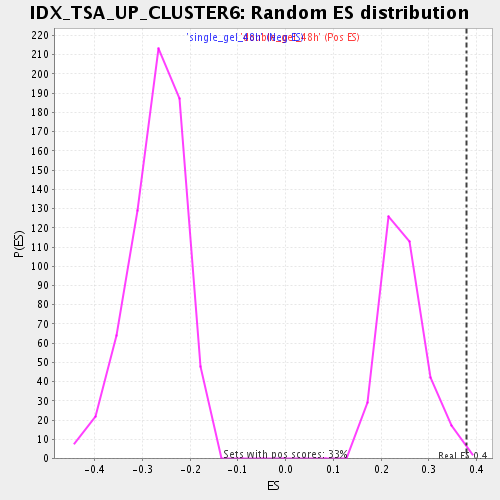
| GeneSet | IDX_TSA_UP_CLUSTER6 |
| Enrichment Score (ES) | 0.378189 |
| Normalized Enrichment Score (NES) | 1.5472419 |
| Nominal p-value | 0.0060790274 |
| FDR q-value | 0.12426227 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FASN | FASN Entrez, Source | fatty acid synthase | 60 | 0.267 | 0.0404 | Yes |
| 2 | ABCA1 | ABCA1 Entrez, Source | ATP-binding cassette, sub-family A (ABC1), member 1 | 71 | 0.251 | 0.0818 | Yes |
| 3 | G0S2 | G0S2 Entrez, Source | G0/G1switch 2 | 98 | 0.228 | 0.1181 | Yes |
| 4 | ACAA2 | ACAA2 Entrez, Source | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | 163 | 0.187 | 0.1448 | Yes |
| 5 | DHCR7 | DHCR7 Entrez, Source | 7-dehydrocholesterol reductase | 247 | 0.160 | 0.1654 | Yes |
| 6 | ALAS1 | ALAS1 Entrez, Source | aminolevulinate, delta-, synthase 1 | 300 | 0.149 | 0.1866 | Yes |
| 7 | HSD11B1 | HSD11B1 Entrez, Source | hydroxysteroid (11-beta) dehydrogenase 1 | 318 | 0.147 | 0.2100 | Yes |
| 8 | CAT | CAT Entrez, Source | catalase | 391 | 0.137 | 0.2276 | Yes |
| 9 | TST | TST Entrez, Source | thiosulfate sulfurtransferase (rhodanese) | 446 | 0.130 | 0.2453 | Yes |
| 10 | FAH | FAH Entrez, Source | fumarylacetoacetate hydrolase (fumarylacetoacetase) | 489 | 0.126 | 0.2633 | Yes |
| 11 | CYB5B | CYB5B Entrez, Source | cytochrome b5 type B (outer mitochondrial membrane) | 498 | 0.125 | 0.2837 | Yes |
| 12 | GPD1 | GPD1 Entrez, Source | glycerol-3-phosphate dehydrogenase 1 (soluble) | 555 | 0.121 | 0.2999 | Yes |
| 13 | REEP6 | REEP6 Entrez, Source | receptor accessory protein 6 | 704 | 0.112 | 0.3075 | Yes |
| 14 | CHPT1 | CHPT1 Entrez, Source | choline phosphotransferase 1 | 789 | 0.107 | 0.3191 | Yes |
| 15 | SLC25A10 | SLC25A10 Entrez, Source | solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 | 790 | 0.107 | 0.3372 | Yes |
| 16 | SLC2A4 | SLC2A4 Entrez, Source | solute carrier family 2 (facilitated glucose transporter), member 4 | 900 | 0.102 | 0.3461 | Yes |
| 17 | NR1H3 | NR1H3 Entrez, Source | nuclear receptor subfamily 1, group H, member 3 | 1041 | 0.096 | 0.3516 | Yes |
| 18 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 1159 | 0.091 | 0.3581 | Yes |
| 19 | PEX19 | PEX19 Entrez, Source | peroxisomal biogenesis factor 19 | 1291 | 0.086 | 0.3627 | Yes |
| 20 | GSR | GSR Entrez, Source | glutathione reductase | 1359 | 0.084 | 0.3717 | Yes |
| 21 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 1493 | 0.080 | 0.3750 | Yes |
| 22 | CIB2 | CIB2 Entrez, Source | calcium and integrin binding family member 2 | 1622 | 0.077 | 0.3782 | Yes |
| 23 | QDPR | QDPR Entrez, Source | quinoid dihydropteridine reductase | 1967 | 0.069 | 0.3638 | No |
| 24 | ETFDH | ETFDH Entrez, Source | electron-transferring-flavoprotein dehydrogenase | 2061 | 0.067 | 0.3679 | No |
| 25 | HBLD2 | HBLD2 Entrez, Source | HESB like domain containing 2 | 2253 | 0.063 | 0.3641 | No |
| 26 | ACAD9 | ACAD9 Entrez, Source | acyl-Coenzyme A dehydrogenase family, member 9 | 2362 | 0.061 | 0.3662 | No |
| 27 | HIBCH | HIBCH Entrez, Source | 3-hydroxyisobutyryl-Coenzyme A hydrolase | 2442 | 0.060 | 0.3702 | No |
| 28 | ALAD | ALAD Entrez, Source | aminolevulinate, delta-, dehydratase | 2787 | 0.054 | 0.3532 | No |
| 29 | HP | HP Entrez, Source | haptoglobin | 2829 | 0.053 | 0.3590 | No |
| 30 | ATP5O | ATP5O Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit (oligomycin sensitivity conferring protein) | 2883 | 0.052 | 0.3637 | No |
| 31 | LIAS | LIAS Entrez, Source | lipoic acid synthetase | 2984 | 0.050 | 0.3646 | No |
| 32 | GBE1 | GBE1 Entrez, Source | glucan (1,4-alpha-), branching enzyme 1 (glycogen branching enzyme, Andersen disease, glycogen storage disease type IV) | 2997 | 0.050 | 0.3721 | No |
| 33 | COL15A1 | COL15A1 Entrez, Source | collagen, type XV, alpha 1 | 3242 | 0.047 | 0.3615 | No |
| 34 | COX7B | COX7B Entrez, Source | cytochrome c oxidase subunit VIIb | 3441 | 0.044 | 0.3539 | No |
| 35 | SLC25A1 | SLC25A1 Entrez, Source | solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 | 3477 | 0.043 | 0.3585 | No |
| 36 | SCP2 | SCP2 Entrez, Source | sterol carrier protein 2 | 3534 | 0.042 | 0.3614 | No |
| 37 | HIBADH | HIBADH Entrez, Source | 3-hydroxyisobutyrate dehydrogenase | 3797 | 0.039 | 0.3482 | No |
| 38 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 3912 | 0.038 | 0.3460 | No |
| 39 | SLC25A5 | SLC25A5 Entrez, Source | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 | 3958 | 0.037 | 0.3488 | No |
| 40 | CAMP | CAMP Entrez, Source | cathelicidin antimicrobial peptide | 4148 | 0.034 | 0.3403 | No |
| 41 | RARRES2 | RARRES2 Entrez, Source | retinoic acid receptor responder (tazarotene induced) 2 | 4262 | 0.033 | 0.3373 | No |
| 42 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 4313 | 0.032 | 0.3390 | No |
| 43 | HSD17B12 | HSD17B12 Entrez, Source | hydroxysteroid (17-beta) dehydrogenase 12 | 4319 | 0.032 | 0.3440 | No |
| 44 | SLC19A1 | SLC19A1 Entrez, Source | solute carrier family 19 (folate transporter), member 1 | 4410 | 0.031 | 0.3425 | No |
| 45 | S100A1 | S100A1 Entrez, Source | S100 calcium binding protein A1 | 4459 | 0.030 | 0.3440 | No |
| 46 | MGST1 | MGST1 Entrez, Source | microsomal glutathione S-transferase 1 | 4492 | 0.030 | 0.3466 | No |
| 47 | AQP7 | AQP7 Entrez, Source | aquaporin 7 | 4534 | 0.030 | 0.3485 | No |
| 48 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 4559 | 0.029 | 0.3516 | No |
| 49 | UQCRC2 | UQCRC2 Entrez, Source | ubiquinol-cytochrome c reductase core protein II | 5022 | 0.024 | 0.3206 | No |
| 50 | MCCC1 | MCCC1 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 1 (alpha) | 5123 | 0.022 | 0.3168 | No |
| 51 | ADIPOQ | ADIPOQ Entrez, Source | adiponectin, C1Q and collagen domain containing | 5211 | 0.021 | 0.3138 | No |
| 52 | PLA2G6 | PLA2G6 Entrez, Source | phospholipase A2, group VI (cytosolic, calcium-independent) | 5243 | 0.021 | 0.3150 | No |
| 53 | TKT | TKT Entrez, Source | transketolase (Wernicke-Korsakoff syndrome) | 5330 | 0.020 | 0.3119 | No |
| 54 | MGLL | MGLL Entrez, Source | monoglyceride lipase | 5400 | 0.019 | 0.3099 | No |
| 55 | NDUFS1 | NDUFS1 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) | 5421 | 0.019 | 0.3116 | No |
| 56 | ORM1 | ORM1 Entrez, Source | orosomucoid 1 | 5486 | 0.019 | 0.3099 | No |
| 57 | SDHC | SDHC Entrez, Source | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | 5522 | 0.018 | 0.3103 | No |
| 58 | EHD1 | EHD1 Entrez, Source | EH-domain containing 1 | 5679 | 0.016 | 0.3012 | No |
| 59 | DLD | DLD Entrez, Source | dihydrolipoamide dehydrogenase | 5876 | 0.014 | 0.2888 | No |
| 60 | DLST | DLST Entrez, Source | dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) | 5882 | 0.014 | 0.2908 | No |
| 61 | COX7A2 | COX7A2 Entrez, Source | cytochrome c oxidase subunit VIIa polypeptide 2 (liver) | 5968 | 0.013 | 0.2866 | No |
| 62 | MOSC2 | MOSC2 Entrez, Source | MOCO sulphurase C-terminal domain containing 2 | 6018 | 0.013 | 0.2850 | No |
| 63 | NDUFA9 | NDUFA9 Entrez, Source | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa | 6270 | 0.010 | 0.2677 | No |
| 64 | MPDU1 | MPDU1 Entrez, Source | mannose-P-dolichol utilization defect 1 | 6280 | 0.010 | 0.2687 | No |
| 65 | UQCRC1 | UQCRC1 Entrez, Source | ubiquinol-cytochrome c reductase core protein I | 6394 | 0.009 | 0.2616 | No |
| 66 | SFXN1 | SFXN1 Entrez, Source | sideroflexin 1 | 6402 | 0.009 | 0.2625 | No |
| 67 | PON2 | PON2 Entrez, Source | paraoxonase 2 | 6716 | 0.005 | 0.2397 | No |
| 68 | DECR1 | DECR1 Entrez, Source | 2,4-dienoyl CoA reductase 1, mitochondrial | 6746 | 0.005 | 0.2383 | No |
| 69 | COX5B | COX5B Entrez, Source | cytochrome c oxidase subunit Vb | 6794 | 0.004 | 0.2355 | No |
| 70 | SDPR | SDPR Entrez, Source | serum deprivation response (phosphatidylserine binding protein) | 6834 | 0.004 | 0.2332 | No |
| 71 | CA2 | CA2 Entrez, Source | carbonic anhydrase II | 7034 | 0.002 | 0.2184 | No |
| 72 | RETSAT | RETSAT Entrez, Source | retinol saturase (all-trans-retinol 13,14-reductase) | 7090 | 0.001 | 0.2145 | No |
| 73 | CDKN2C | CDKN2C Entrez, Source | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | 7160 | 0.000 | 0.2093 | No |
| 74 | PGM1 | PGM1 Entrez, Source | phosphoglucomutase 1 | 7335 | -0.002 | 0.1964 | No |
| 75 | CYB5A | CYB5A Entrez, Source | cytochrome b5 type A (microsomal) | 7424 | -0.003 | 0.1903 | No |
| 76 | ATP5D | ATP5D Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit | 7712 | -0.006 | 0.1696 | No |
| 77 | HADH | HADH Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase | 7824 | -0.007 | 0.1624 | No |
| 78 | CS | CS Entrez, Source | citrate synthase | 7918 | -0.009 | 0.1568 | No |
| 79 | PDAP1 | PDAP1 Entrez, Source | PDGFA associated protein 1 | 7929 | -0.009 | 0.1575 | No |
| 80 | LTC4S | LTC4S Entrez, Source | leukotriene C4 synthase | 7935 | -0.009 | 0.1586 | No |
| 81 | ACADS | ACADS Entrez, Source | acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain | 7949 | -0.009 | 0.1591 | No |
| 82 | UQCRB | UQCRB Entrez, Source | ubiquinol-cytochrome c reductase binding protein | 8079 | -0.010 | 0.1511 | No |
| 83 | ISOC1 | ISOC1 Entrez, Source | isochorismatase domain containing 1 | 8281 | -0.013 | 0.1381 | No |
| 84 | SUCLG1 | SUCLG1 Entrez, Source | succinate-CoA ligase, GDP-forming, alpha subunit | 8338 | -0.013 | 0.1361 | No |
| 85 | SAMM50 | SAMM50 Entrez, Source | sorting and assembly machinery component 50 homolog (S. cerevisiae) | 8343 | -0.013 | 0.1381 | No |
| 86 | NDUFA5 | NDUFA5 Entrez, Source | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa | 8599 | -0.017 | 0.1216 | No |
| 87 | BNIP3 | BNIP3 Entrez, Source | BCL2/adenovirus E1B 19kDa interacting protein 3 | 8690 | -0.018 | 0.1178 | No |
| 88 | LPL | LPL Entrez, Source | lipoprotein lipase | 8894 | -0.020 | 0.1059 | No |
| 89 | DBT | DBT Entrez, Source | dihydrolipoamide branched chain transacylase E2 | 8987 | -0.022 | 0.1026 | No |
| 90 | PCAF | PCAF Entrez, Source | p300/CBP-associated factor | 9640 | -0.031 | 0.0586 | No |
| 91 | ACADL | ACADL Entrez, Source | acyl-Coenzyme A dehydrogenase, long chain | 9656 | -0.031 | 0.0627 | No |
| 92 | SCARB1 | SCARB1 Entrez, Source | scavenger receptor class B, member 1 | 9712 | -0.032 | 0.0640 | No |
| 93 | DGAT1 | DGAT1 Entrez, Source | diacylglycerol O-acyltransferase homolog 1 (mouse) | 10079 | -0.038 | 0.0427 | No |
| 94 | TSPAN12 | TSPAN12 Entrez, Source | tetraspanin 12 | 10187 | -0.040 | 0.0413 | No |
| 95 | ABCD2 | ABCD2 Entrez, Source | ATP-binding cassette, sub-family D (ALD), member 2 | 10245 | -0.041 | 0.0438 | No |
| 96 | BCAT2 | BCAT2 Entrez, Source | branched chain aminotransferase 2, mitochondrial | 10290 | -0.041 | 0.0474 | No |
| 97 | RTN3 | RTN3 Entrez, Source | reticulon 3 | 10382 | -0.043 | 0.0478 | No |
| 98 | CBR1 | CBR1 Entrez, Source | carbonyl reductase 1 | 10804 | -0.052 | 0.0246 | No |
| 99 | ADCY6 | ADCY6 Entrez, Source | adenylate cyclase 6 | 10943 | -0.055 | 0.0234 | No |
| 100 | S100A8 | S100A8 Entrez, Source | S100 calcium binding protein A8 | 11047 | -0.057 | 0.0252 | No |
| 101 | DHRS7B | DHRS7B Entrez, Source | dehydrogenase/reductase (SDR family) member 7B | 11284 | -0.062 | 0.0177 | No |
| 102 | PSEN2 | PSEN2 Entrez, Source | presenilin 2 (Alzheimer disease 4) | 11726 | -0.076 | -0.0028 | No |
| 103 | PCYOX1 | PCYOX1 Entrez, Source | prenylcysteine oxidase 1 | 11731 | -0.077 | 0.0098 | No |
| 104 | UNC119 | UNC119 Entrez, Source | unc-119 homolog (C. elegans) | 11827 | -0.081 | 0.0161 | No |
| 105 | DLAT | DLAT Entrez, Source | dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) | 12499 | -0.117 | -0.0149 | No |
| 106 | RAB3D | RAB3D Entrez, Source | RAB3D, member RAS oncogene family | 12543 | -0.121 | 0.0022 | No |
| 107 | ITGA6 | ITGA6 Entrez, Source | integrin, alpha 6 | 12716 | -0.137 | 0.0122 | No |
| 108 | LDHB | LDHB Entrez, Source | lactate dehydrogenase B | 13053 | -0.209 | 0.0218 | No |