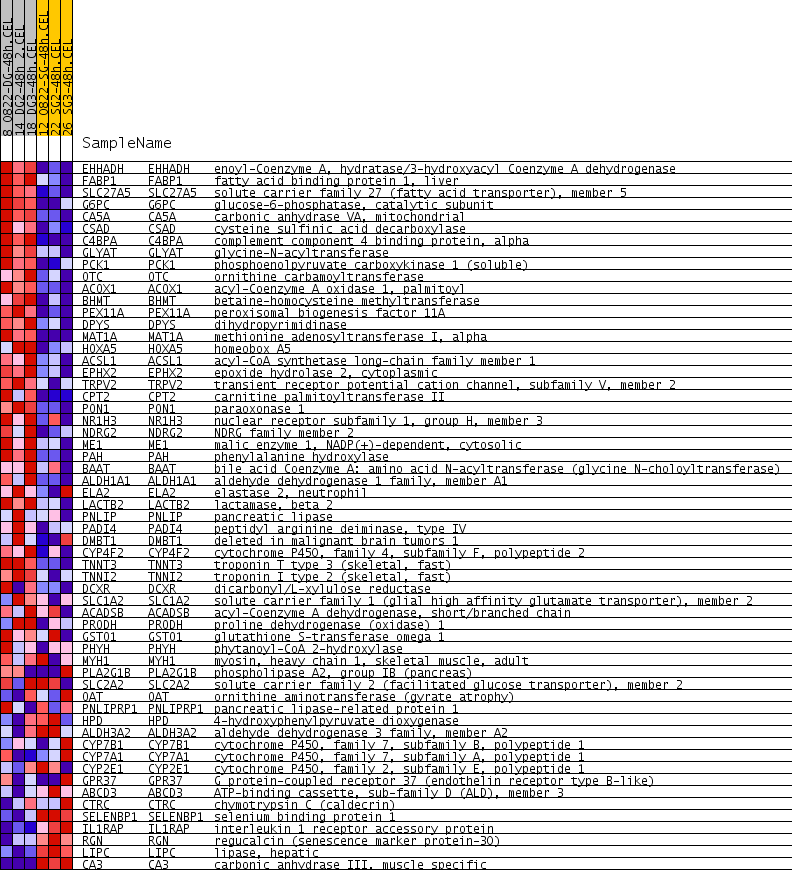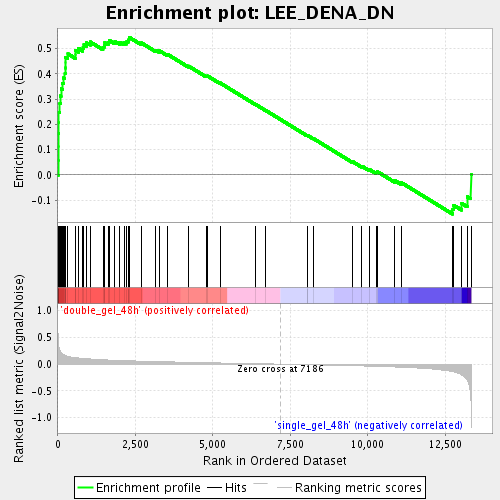
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | LEE_DENA_DN |
| Enrichment Score (ES) | 0.54510075 |
| Normalized Enrichment Score (NES) | 2.0049574 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0060135396 |
| FWER p-Value | 0.177 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 6 | 0.447 | 0.0587 | Yes |
| 2 | FABP1 | FABP1 Entrez, Source | fatty acid binding protein 1, liver | 10 | 0.411 | 0.1128 | Yes |
| 3 | SLC27A5 | SLC27A5 Entrez, Source | solute carrier family 27 (fatty acid transporter), member 5 | 12 | 0.395 | 0.1650 | Yes |
| 4 | G6PC | G6PC Entrez, Source | glucose-6-phosphatase, catalytic subunit | 26 | 0.337 | 0.2087 | Yes |
| 5 | CA5A | CA5A Entrez, Source | carbonic anhydrase VA, mitochondrial | 32 | 0.310 | 0.2492 | Yes |
| 6 | CSAD | CSAD Entrez, Source | cysteine sulfinic acid decarboxylase | 62 | 0.265 | 0.2821 | Yes |
| 7 | C4BPA | C4BPA Entrez, Source | complement component 4 binding protein, alpha | 79 | 0.243 | 0.3131 | Yes |
| 8 | GLYAT | GLYAT Entrez, Source | glycine-N-acyltransferase | 102 | 0.225 | 0.3411 | Yes |
| 9 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 148 | 0.193 | 0.3632 | Yes |
| 10 | OTC | OTC Entrez, Source | ornithine carbamoyltransferase | 183 | 0.178 | 0.3842 | Yes |
| 11 | ACOX1 | ACOX1 Entrez, Source | acyl-Coenzyme A oxidase 1, palmitoyl | 227 | 0.165 | 0.4028 | Yes |
| 12 | BHMT | BHMT Entrez, Source | betaine-homocysteine methyltransferase | 237 | 0.162 | 0.4235 | Yes |
| 13 | PEX11A | PEX11A Entrez, Source | peroxisomal biogenesis factor 11A | 241 | 0.161 | 0.4446 | Yes |
| 14 | DPYS | DPYS Entrez, Source | dihydropyrimidinase | 242 | 0.161 | 0.4659 | Yes |
| 15 | MAT1A | MAT1A Entrez, Source | methionine adenosyltransferase I, alpha | 323 | 0.146 | 0.4792 | Yes |
| 16 | HOXA5 | HOXA5 Entrez, Source | homeobox A5 | 562 | 0.121 | 0.4773 | Yes |
| 17 | ACSL1 | ACSL1 Entrez, Source | acyl-CoA synthetase long-chain family member 1 | 564 | 0.121 | 0.4931 | Yes |
| 18 | EPHX2 | EPHX2 Entrez, Source | epoxide hydrolase 2, cytoplasmic | 660 | 0.114 | 0.5011 | Yes |
| 19 | TRPV2 | TRPV2 Entrez, Source | transient receptor potential cation channel, subfamily V, member 2 | 808 | 0.106 | 0.5041 | Yes |
| 20 | CPT2 | CPT2 Entrez, Source | carnitine palmitoyltransferase II | 840 | 0.105 | 0.5157 | Yes |
| 21 | PON1 | PON1 Entrez, Source | paraoxonase 1 | 922 | 0.101 | 0.5230 | Yes |
| 22 | NR1H3 | NR1H3 Entrez, Source | nuclear receptor subfamily 1, group H, member 3 | 1041 | 0.096 | 0.5268 | Yes |
| 23 | NDRG2 | NDRG2 Entrez, Source | NDRG family member 2 | 1456 | 0.081 | 0.5063 | Yes |
| 24 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 1493 | 0.080 | 0.5141 | Yes |
| 25 | PAH | PAH Entrez, Source | phenylalanine hydroxylase | 1513 | 0.079 | 0.5231 | Yes |
| 26 | BAAT | BAAT Entrez, Source | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 1625 | 0.077 | 0.5249 | Yes |
| 27 | ALDH1A1 | ALDH1A1 Entrez, Source | aldehyde dehydrogenase 1 family, member A1 | 1653 | 0.076 | 0.5329 | Yes |
| 28 | ELA2 | ELA2 Entrez, Source | elastase 2, neutrophil | 1838 | 0.071 | 0.5285 | Yes |
| 29 | LACTB2 | LACTB2 Entrez, Source | lactamase, beta 2 | 1997 | 0.068 | 0.5256 | Yes |
| 30 | PNLIP | PNLIP Entrez, Source | pancreatic lipase | 2144 | 0.065 | 0.5232 | Yes |
| 31 | PADI4 | PADI4 Entrez, Source | peptidyl arginine deiminase, type IV | 2211 | 0.064 | 0.5267 | Yes |
| 32 | DMBT1 | DMBT1 Entrez, Source | deleted in malignant brain tumors 1 | 2265 | 0.063 | 0.5310 | Yes |
| 33 | CYP4F2 | CYP4F2 Entrez, Source | cytochrome P450, family 4, subfamily F, polypeptide 2 | 2291 | 0.062 | 0.5373 | Yes |
| 34 | TNNT3 | TNNT3 Entrez, Source | troponin T type 3 (skeletal, fast) | 2298 | 0.062 | 0.5451 | Yes |
| 35 | TNNI2 | TNNI2 Entrez, Source | troponin I type 2 (skeletal, fast) | 2692 | 0.055 | 0.5228 | No |
| 36 | DCXR | DCXR Entrez, Source | dicarbonyl/L-xylulose reductase | 3163 | 0.048 | 0.4938 | No |
| 37 | SLC1A2 | SLC1A2 Entrez, Source | solute carrier family 1 (glial high affinity glutamate transporter), member 2 | 3274 | 0.046 | 0.4916 | No |
| 38 | ACADSB | ACADSB Entrez, Source | acyl-Coenzyme A dehydrogenase, short/branched chain | 3526 | 0.043 | 0.4783 | No |
| 39 | PRODH | PRODH Entrez, Source | proline dehydrogenase (oxidase) 1 | 4210 | 0.034 | 0.4313 | No |
| 40 | GSTO1 | GSTO1 Entrez, Source | glutathione S-transferase omega 1 | 4785 | 0.026 | 0.3916 | No |
| 41 | PHYH | PHYH Entrez, Source | phytanoyl-CoA 2-hydroxylase | 4827 | 0.026 | 0.3920 | No |
| 42 | MYH1 | MYH1 Entrez, Source | myosin, heavy chain 1, skeletal muscle, adult | 5231 | 0.021 | 0.3645 | No |
| 43 | PLA2G1B | PLA2G1B Entrez, Source | phospholipase A2, group IB (pancreas) | 6366 | 0.009 | 0.2803 | No |
| 44 | SLC2A2 | SLC2A2 Entrez, Source | solute carrier family 2 (facilitated glucose transporter), member 2 | 6701 | 0.005 | 0.2558 | No |
| 45 | OAT | OAT Entrez, Source | ornithine aminotransferase (gyrate atrophy) | 8046 | -0.010 | 0.1560 | No |
| 46 | PNLIPRP1 | PNLIPRP1 Entrez, Source | pancreatic lipase-related protein 1 | 8063 | -0.010 | 0.1561 | No |
| 47 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 8248 | -0.012 | 0.1439 | No |
| 48 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 9511 | -0.029 | 0.0527 | No |
| 49 | CYP7B1 | CYP7B1 Entrez, Source | cytochrome P450, family 7, subfamily B, polypeptide 1 | 9803 | -0.033 | 0.0353 | No |
| 50 | CYP7A1 | CYP7A1 Entrez, Source | cytochrome P450, family 7, subfamily A, polypeptide 1 | 10060 | -0.038 | 0.0210 | No |
| 51 | CYP2E1 | CYP2E1 Entrez, Source | cytochrome P450, family 2, subfamily E, polypeptide 1 | 10268 | -0.041 | 0.0108 | No |
| 52 | GPR37 | GPR37 Entrez, Source | G protein-coupled receptor 37 (endothelin receptor type B-like) | 10305 | -0.041 | 0.0136 | No |
| 53 | ABCD3 | ABCD3 Entrez, Source | ATP-binding cassette, sub-family D (ALD), member 3 | 10872 | -0.053 | -0.0220 | No |
| 54 | CTRC | CTRC Entrez, Source | chymotrypsin C (caldecrin) | 11080 | -0.058 | -0.0300 | No |
| 55 | SELENBP1 | SELENBP1 Entrez, Source | selenium binding protein 1 | 12740 | -0.140 | -0.1364 | No |
| 56 | IL1RAP | IL1RAP Entrez, Source | interleukin 1 receptor accessory protein | 12771 | -0.144 | -0.1196 | No |
| 57 | RGN | RGN Entrez, Source | regucalcin (senescence marker protein-30) | 13030 | -0.199 | -0.1127 | No |
| 58 | LIPC | LIPC Entrez, Source | lipase, hepatic | 13211 | -0.304 | -0.0861 | No |
| 59 | CA3 | CA3 Entrez, Source | carbonic anhydrase III, muscle specific | 13332 | -0.725 | 0.0008 | No |