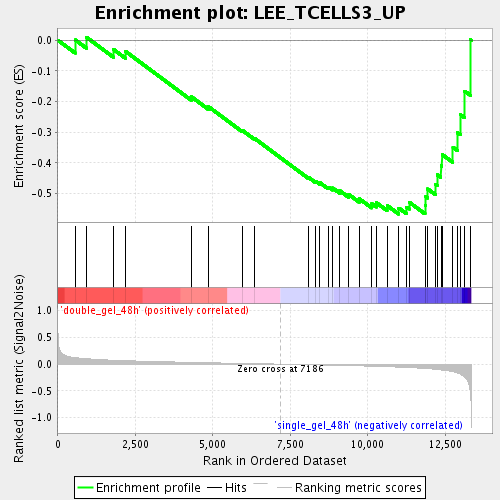
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | single_gel_48h |
| GeneSet | LEE_TCELLS3_UP |
| Enrichment Score (ES) | -0.56700724 |
| Normalized Enrichment Score (NES) | -1.7272772 |
| Nominal p-value | 0.0072332732 |
| FDR q-value | 0.055199854 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TSHR | TSHR Entrez, Source | thyroid stimulating hormone receptor | 557 | 0.121 | 0.0012 | No |
| 2 | PON1 | PON1 Entrez, Source | paraoxonase 1 | 922 | 0.101 | 0.0097 | No |
| 3 | RAPGEF5 | RAPGEF5 Entrez, Source | Rap guanine nucleotide exchange factor (GEF) 5 | 1793 | 0.072 | -0.0299 | No |
| 4 | GALNT7 | GALNT7 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) | 2182 | 0.064 | -0.0363 | No |
| 5 | CDKN3 | CDKN3 Entrez, Source | cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | 4303 | 0.033 | -0.1840 | No |
| 6 | CDC25A | CDC25A Entrez, Source | cell division cycle 25A | 4860 | 0.026 | -0.2167 | No |
| 7 | SYK | SYK Entrez, Source | spleen tyrosine kinase | 5948 | 0.013 | -0.2936 | No |
| 8 | LETM1 | LETM1 Entrez, Source | leucine zipper-EF-hand containing transmembrane protein 1 | 6342 | 0.009 | -0.3199 | No |
| 9 | SLC1A4 | SLC1A4 Entrez, Source | solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 | 8089 | -0.010 | -0.4474 | No |
| 10 | GNA15 | GNA15 Entrez, Source | guanine nucleotide binding protein (G protein), alpha 15 (Gq class) | 8319 | -0.013 | -0.4599 | No |
| 11 | E2F8 | E2F8 Entrez, Source | E2F transcription factor 8 | 8457 | -0.015 | -0.4648 | No |
| 12 | CDCA1 | CDCA1 Entrez, Source | cell division cycle associated 1 | 8728 | -0.018 | -0.4786 | No |
| 13 | ADA | ADA Entrez, Source | adenosine deaminase | 8863 | -0.020 | -0.4816 | No |
| 14 | TUBB2C | TUBB2C Entrez, Source | tubulin, beta 2C | 9095 | -0.023 | -0.4908 | No |
| 15 | CXXC5 | CXXC5 Entrez, Source | CXXC finger 5 | 9385 | -0.027 | -0.5028 | No |
| 16 | AURKA | AURKA Entrez, Source | aurora kinase A | 9722 | -0.032 | -0.5165 | No |
| 17 | TOP2A | TOP2A Entrez, Source | topoisomerase (DNA) II alpha 170kDa | 10131 | -0.039 | -0.5334 | No |
| 18 | GFI1 | GFI1 Entrez, Source | growth factor independent 1 | 10274 | -0.041 | -0.5295 | No |
| 19 | PTTG1 | PTTG1 Entrez, Source | pituitary tumor-transforming 1 | 10625 | -0.047 | -0.5390 | No |
| 20 | CDCA2 | CDCA2 Entrez, Source | cell division cycle associated 2 | 10999 | -0.056 | -0.5472 | Yes |
| 21 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 11253 | -0.061 | -0.5444 | Yes |
| 22 | BUB1B | BUB1B Entrez, Source | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | 11337 | -0.063 | -0.5281 | Yes |
| 23 | HMMR | HMMR Entrez, Source | hyaluronan-mediated motility receptor (RHAMM) | 11850 | -0.081 | -0.5378 | Yes |
| 24 | CD99 | CD99 Entrez, Source | CD99 molecule | 11871 | -0.082 | -0.5102 | Yes |
| 25 | FAIM | FAIM Entrez, Source | Fas apoptotic inhibitory molecule | 11934 | -0.084 | -0.4849 | Yes |
| 26 | DSCR1 | DSCR1 Entrez, Source | Down syndrome critical region gene 1 | 12190 | -0.096 | -0.4699 | Yes |
| 27 | GGH | GGH Entrez, Source | gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) | 12243 | -0.099 | -0.4386 | Yes |
| 28 | TUBB | TUBB Entrez, Source | tubulin, beta | 12364 | -0.108 | -0.4092 | Yes |
| 29 | CCNB2 | CCNB2 Entrez, Source | cyclin B2 | 12396 | -0.111 | -0.3723 | Yes |
| 30 | TYMS | TYMS Entrez, Source | thymidylate synthetase | 12749 | -0.141 | -0.3488 | Yes |
| 31 | KIF2C | KIF2C Entrez, Source | kinesin family member 2C | 12895 | -0.164 | -0.3016 | Yes |
| 32 | SCRN1 | SCRN1 Entrez, Source | secernin 1 | 12993 | -0.186 | -0.2427 | Yes |
| 33 | CCNB1 | CCNB1 Entrez, Source | cyclin B1 | 13126 | -0.245 | -0.1656 | Yes |
| 34 | KIAA0101 | KIAA0101 Entrez, Source | KIAA0101 | 13316 | -0.512 | 0.0020 | Yes |

