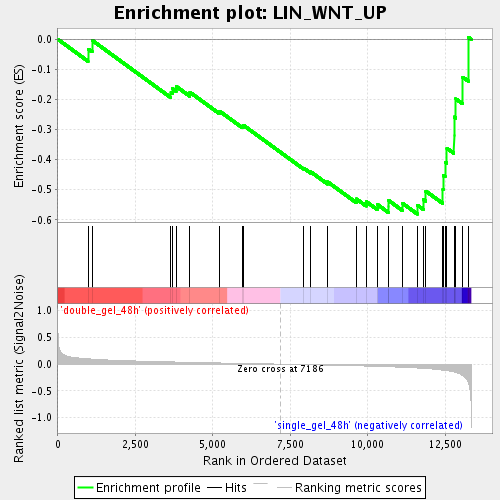
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | single_gel_48h |
| GeneSet | LIN_WNT_UP |
| Enrichment Score (ES) | -0.58107704 |
| Normalized Enrichment Score (NES) | -1.7032573 |
| Nominal p-value | 0.0017667845 |
| FDR q-value | 0.06587836 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ATP6V0A1 | ATP6V0A1 Entrez, Source | ATPase, H+ transporting, lysosomal V0 subunit a1 | 974 | 0.099 | -0.0322 | No |
| 2 | GJB1 | GJB1 Entrez, Source | gap junction protein, beta 1, 32kDa (connexin 32, Charcot-Marie-Tooth neuropathy, X-linked) | 1107 | 0.093 | -0.0034 | No |
| 3 | IMPDH2 | IMPDH2 Entrez, Source | IMP (inosine monophosphate) dehydrogenase 2 | 3648 | 0.041 | -0.1770 | No |
| 4 | SEC61A1 | SEC61A1 Entrez, Source | Sec61 alpha 1 subunit (S. cerevisiae) | 3698 | 0.040 | -0.1639 | No |
| 5 | MTA1 | MTA1 Entrez, Source | metastasis associated 1 | 3816 | 0.039 | -0.1564 | No |
| 6 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 4258 | 0.033 | -0.1758 | No |
| 7 | HSPE1 | HSPE1 Entrez, Source | heat shock 10kDa protein 1 (chaperonin 10) | 5218 | 0.021 | -0.2390 | No |
| 8 | SYK | SYK Entrez, Source | spleen tyrosine kinase | 5948 | 0.013 | -0.2882 | No |
| 9 | EIF5A | EIF5A Entrez, Source | eukaryotic translation initiation factor 5A | 5996 | 0.013 | -0.2864 | No |
| 10 | PDAP1 | PDAP1 Entrez, Source | PDGFA associated protein 1 | 7929 | -0.009 | -0.4279 | No |
| 11 | ARHGDIA | ARHGDIA Entrez, Source | Rho GDP dissociation inhibitor (GDI) alpha | 8158 | -0.011 | -0.4404 | No |
| 12 | ALG3 | ALG3 Entrez, Source | asparagine-linked glycosylation 3 homolog (S. cerevisiae, alpha-1,3-mannosyltransferase) | 8704 | -0.018 | -0.4738 | No |
| 13 | HDGF | HDGF Entrez, Source | hepatoma-derived growth factor (high-mobility group protein 1-like) | 9633 | -0.031 | -0.5306 | No |
| 14 | BCAP31 | BCAP31 Entrez, Source | B-cell receptor-associated protein 31 | 9950 | -0.036 | -0.5395 | No |
| 15 | PFN1 | PFN1 Entrez, Source | profilin 1 | 10319 | -0.042 | -0.5498 | No |
| 16 | GLTSCR2 | GLTSCR2 Entrez, Source | glioma tumor suppressor candidate region gene 2 | 10673 | -0.049 | -0.5561 | Yes |
| 17 | MAP3K11 | MAP3K11 Entrez, Source | mitogen-activated protein kinase kinase kinase 11 | 10674 | -0.049 | -0.5359 | Yes |
| 18 | AES | AES Entrez, Source | amino-terminal enhancer of split | 11117 | -0.058 | -0.5448 | Yes |
| 19 | SLC1A5 | SLC1A5 Entrez, Source | solute carrier family 1 (neutral amino acid transporter), member 5 | 11601 | -0.072 | -0.5511 | Yes |
| 20 | MMP14 | MMP14 Entrez, Source | matrix metallopeptidase 14 (membrane-inserted) | 11798 | -0.079 | -0.5331 | Yes |
| 21 | CD99 | CD99 Entrez, Source | CD99 molecule | 11871 | -0.082 | -0.5044 | Yes |
| 22 | CAPN1 | CAPN1 Entrez, Source | calpain 1, (mu/I) large subunit | 12425 | -0.112 | -0.4992 | Yes |
| 23 | GNA12 | GNA12 Entrez, Source | guanine nucleotide binding protein (G protein) alpha 12 | 12436 | -0.113 | -0.4528 | Yes |
| 24 | FLOT2 | FLOT2 Entrez, Source | flotillin 2 | 12500 | -0.117 | -0.4087 | Yes |
| 25 | LMNA | LMNA Entrez, Source | lamin A/C | 12554 | -0.122 | -0.3620 | Yes |
| 26 | TGM2 | TGM2 Entrez, Source | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | 12783 | -0.145 | -0.3187 | Yes |
| 27 | ZBTB7A | ZBTB7A Entrez, Source | zinc finger and BTB domain containing 7A | 12784 | -0.146 | -0.2581 | Yes |
| 28 | ACTN4 | ACTN4 Entrez, Source | actinin, alpha 4 | 12842 | -0.155 | -0.1980 | Yes |
| 29 | ETS1 | ETS1 Entrez, Source | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | 13063 | -0.214 | -0.1254 | Yes |
| 30 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 13250 | -0.352 | 0.0069 | Yes |