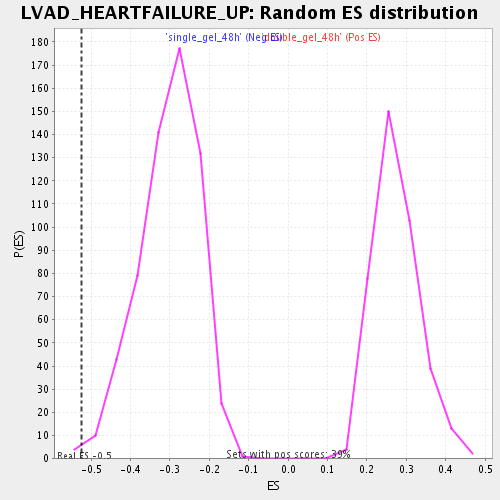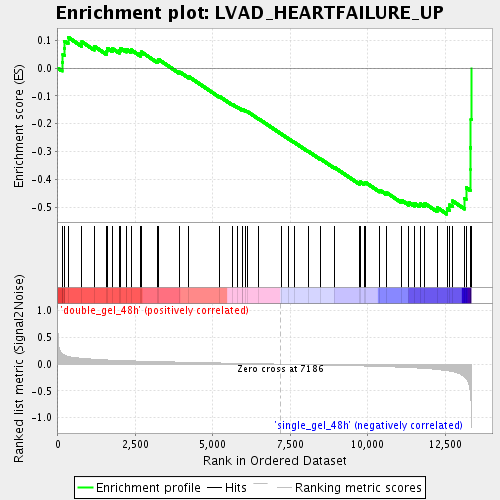
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | single_gel_48h |
| GeneSet | LVAD_HEARTFAILURE_UP |
| Enrichment Score (ES) | -0.5253509 |
| Normalized Enrichment Score (NES) | -1.7446537 |
| Nominal p-value | 0.004909984 |
| FDR q-value | 0.050661184 |
| FWER p-Value | 0.999 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DDIT4 | DDIT4 Entrez, Source | DNA-damage-inducible transcript 4 | 134 | 0.198 | 0.0208 | No |
| 2 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 160 | 0.189 | 0.0484 | No |
| 3 | PDK4 | PDK4 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 4 | 207 | 0.172 | 0.0717 | No |
| 4 | FKBP5 | FKBP5 Entrez, Source | FK506 binding protein 5 | 219 | 0.169 | 0.0971 | No |
| 5 | ATF3 | ATF3 Entrez, Source | activating transcription factor 3 | 329 | 0.145 | 0.1115 | No |
| 6 | INSR | INSR Entrez, Source | insulin receptor | 772 | 0.108 | 0.0950 | No |
| 7 | SLCO4A1 | SLCO4A1 Entrez, Source | solute carrier organic anion transporter family, member 4A1 | 1167 | 0.091 | 0.0795 | No |
| 8 | CHST3 | CHST3 Entrez, Source | carbohydrate (chondroitin 6) sulfotransferase 3 | 1564 | 0.078 | 0.0618 | No |
| 9 | KLF9 | KLF9 Entrez, Source | Kruppel-like factor 9 | 1585 | 0.077 | 0.0724 | No |
| 10 | CDC37L1 | CDC37L1 Entrez, Source | CDC37 cell division cycle 37 homolog (S. cerevisiae)-like 1 | 1754 | 0.073 | 0.0711 | No |
| 11 | AREG | AREG Entrez, Source | amphiregulin (schwannoma-derived growth factor) | 1977 | 0.069 | 0.0651 | No |
| 12 | MYOM1 | MYOM1 Entrez, Source | myomesin 1 (skelemin) 185kDa | 2024 | 0.067 | 0.0721 | No |
| 13 | CSDC2 | CSDC2 Entrez, Source | cold shock domain containing C2, RNA binding | 2209 | 0.064 | 0.0682 | No |
| 14 | FLNC | FLNC Entrez, Source | filamin C, gamma (actin binding protein 280) | 2361 | 0.061 | 0.0663 | No |
| 15 | PIK3R1 | PIK3R1 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 1 (p85 alpha) | 2668 | 0.056 | 0.0520 | No |
| 16 | NFIL3 | NFIL3 Entrez, Source | nuclear factor, interleukin 3 regulated | 2690 | 0.055 | 0.0590 | No |
| 17 | FHL1 | FHL1 Entrez, Source | four and a half LIM domains 1 | 3226 | 0.047 | 0.0261 | No |
| 18 | CYP4B1 | CYP4B1 Entrez, Source | cytochrome P450, family 4, subfamily B, polypeptide 1 | 3238 | 0.047 | 0.0325 | No |
| 19 | BTG2 | BTG2 Entrez, Source | BTG family, member 2 | 3927 | 0.037 | -0.0134 | No |
| 20 | CEBPD | CEBPD Entrez, Source | CCAAT/enhancer binding protein (C/EBP), delta | 4223 | 0.034 | -0.0304 | No |
| 21 | HYAL2 | HYAL2 Entrez, Source | hyaluronoglucosaminidase 2 | 5215 | 0.021 | -0.1017 | No |
| 22 | AMD1 | AMD1 Entrez, Source | adenosylmethionine decarboxylase 1 | 5628 | 0.017 | -0.1301 | No |
| 23 | CDKN1A | CDKN1A Entrez, Source | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 5802 | 0.015 | -0.1408 | No |
| 24 | ADFP | ADFP Entrez, Source | adipose differentiation-related protein | 5942 | 0.013 | -0.1491 | No |
| 25 | PER1 | PER1 Entrez, Source | period homolog 1 (Drosophila) | 5944 | 0.013 | -0.1471 | No |
| 26 | NID1 | NID1 Entrez, Source | nidogen 1 | 6040 | 0.012 | -0.1523 | No |
| 27 | CNN1 | CNN1 Entrez, Source | calponin 1, basic, smooth muscle | 6113 | 0.012 | -0.1559 | No |
| 28 | TSC22D3 | TSC22D3 Entrez, Source | TSC22 domain family, member 3 | 6484 | 0.008 | -0.1826 | No |
| 29 | DOCK9 | DOCK9 Entrez, Source | dedicator of cytokinesis 9 | 7211 | -0.000 | -0.2371 | No |
| 30 | KCNK1 | KCNK1 Entrez, Source | potassium channel, subfamily K, member 1 | 7451 | -0.003 | -0.2546 | No |
| 31 | ZBTB16 | ZBTB16 Entrez, Source | zinc finger and BTB domain containing 16 | 7635 | -0.005 | -0.2676 | No |
| 32 | JUN | JUN Entrez, Source | jun oncogene | 8081 | -0.010 | -0.2995 | No |
| 33 | RERE | RERE Entrez, Source | arginine-glutamic acid dipeptide (RE) repeats | 8473 | -0.015 | -0.3265 | No |
| 34 | RGC32 | RGC32 Entrez, Source | - | 8939 | -0.021 | -0.3582 | No |
| 35 | GLUL | GLUL Entrez, Source | glutamate-ammonia ligase (glutamine synthetase) | 9734 | -0.033 | -0.4129 | No |
| 36 | RND3 | RND3 Entrez, Source | Rho family GTPase 3 | 9751 | -0.033 | -0.4090 | No |
| 37 | AGTR1 | AGTR1 Entrez, Source | angiotensin II receptor, type 1 | 9880 | -0.035 | -0.4133 | No |
| 38 | SLC38A2 | SLC38A2 Entrez, Source | solute carrier family 38, member 2 | 9913 | -0.035 | -0.4102 | No |
| 39 | NFKBIA | NFKBIA Entrez, Source | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | 10388 | -0.043 | -0.4392 | No |
| 40 | GADD45B | GADD45B Entrez, Source | growth arrest and DNA-damage-inducible, beta | 10614 | -0.047 | -0.4487 | No |
| 41 | PPAP2A | PPAP2A Entrez, Source | phosphatidic acid phosphatase type 2A | 11084 | -0.058 | -0.4751 | No |
| 42 | IER5 | IER5 Entrez, Source | immediate early response 5 | 11328 | -0.063 | -0.4835 | No |
| 43 | MFAP4 | MFAP4 Entrez, Source | microfibrillar-associated protein 4 | 11503 | -0.068 | -0.4860 | No |
| 44 | CLU | CLU Entrez, Source | clusterin | 11686 | -0.075 | -0.4880 | No |
| 45 | FSTL3 | FSTL3 Entrez, Source | follistatin-like 3 (secreted glycoprotein) | 11840 | -0.081 | -0.4869 | No |
| 46 | DIXDC1 | DIXDC1 Entrez, Source | DIX domain containing 1 | 12239 | -0.099 | -0.5015 | No |
| 47 | CYR61 | CYR61 Entrez, Source | cysteine-rich, angiogenic inducer, 61 | 12557 | -0.122 | -0.5064 | Yes |
| 48 | ELF1 | ELF1 Entrez, Source | E74-like factor 1 (ets domain transcription factor) | 12637 | -0.129 | -0.4923 | Yes |
| 49 | CALD1 | CALD1 Entrez, Source | caldesmon 1 | 12735 | -0.139 | -0.4779 | Yes |
| 50 | RRAS2 | RRAS2 Entrez, Source | related RAS viral (r-ras) oncogene homolog 2 | 13125 | -0.244 | -0.4691 | Yes |
| 51 | TIMP3 | TIMP3 Entrez, Source | TIMP metallopeptidase inhibitor 3 (Sorsby fundus dystrophy, pseudoinflammatory) | 13175 | -0.275 | -0.4300 | Yes |
| 52 | SERPINE1 | SERPINE1 Entrez, Source | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 13308 | -0.484 | -0.3645 | Yes |
| 53 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 13313 | -0.503 | -0.2865 | Yes |
| 54 | ANKRD1 | ANKRD1 Entrez, Source | ankyrin repeat domain 1 (cardiac muscle) | 13330 | -0.666 | -0.1840 | Yes |
| 55 | ACTA1 | ACTA1 Entrez, Source | actin, alpha 1, skeletal muscle | 13342 | -1.187 | 0.0000 | Yes |