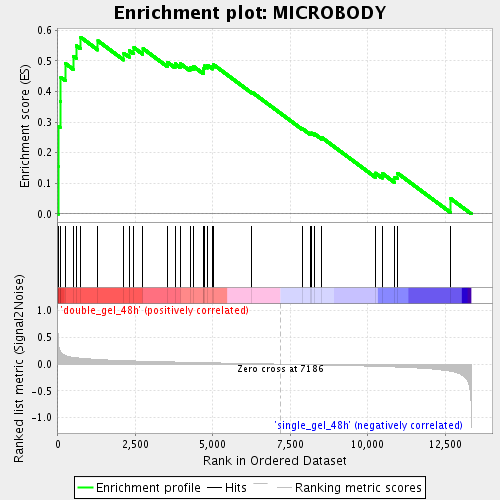
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | MICROBODY |
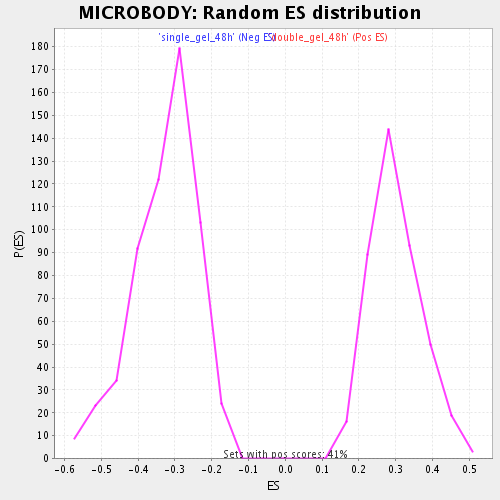
| Enrichment Score (ES) | 0.5769349 |
| Normalized Enrichment Score (NES) | 1.9270296 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.012881649 |
| FWER p-Value | 0.442 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 6 | 0.447 | 0.1546 | Yes |
| 2 | IDI1 | IDI1 Entrez, Source | isopentenyl-diphosphate delta isomerase 1 | 13 | 0.382 | 0.2869 | Yes |
| 3 | PMVK | PMVK Entrez, Source | phosphomevalonate kinase | 73 | 0.247 | 0.3683 | Yes |
| 4 | HACL1 | HACL1 Entrez, Source | 2-hydroxyacyl-CoA lyase 1 | 94 | 0.229 | 0.4463 | Yes |
| 5 | PEX11A | PEX11A Entrez, Source | peroxisomal biogenesis factor 11A | 241 | 0.161 | 0.4912 | Yes |
| 6 | PEX16 | PEX16 Entrez, Source | peroxisomal biogenesis factor 16 | 496 | 0.125 | 0.5156 | Yes |
| 7 | AGXT | AGXT Entrez, Source | alanine-glyoxylate aminotransferase (oxalosis I; hyperoxaluria I; glycolicaciduria; serine-pyruvate aminotransferase) | 591 | 0.118 | 0.5496 | Yes |
| 8 | AMACR | AMACR Entrez, Source | alpha-methylacyl-CoA racemase | 735 | 0.110 | 0.5769 | Yes |
| 9 | PEX19 | PEX19 Entrez, Source | peroxisomal biogenesis factor 19 | 1291 | 0.086 | 0.5651 | No |
| 10 | PXMP4 | PXMP4 Entrez, Source | peroxisomal membrane protein 4, 24kDa | 2125 | 0.065 | 0.5252 | No |
| 11 | MLYCD | MLYCD Entrez, Source | malonyl-CoA decarboxylase | 2302 | 0.062 | 0.5335 | No |
| 12 | PEX12 | PEX12 Entrez, Source | peroxisomal biogenesis factor 12 | 2450 | 0.060 | 0.5431 | No |
| 13 | PEX3 | PEX3 Entrez, Source | peroxisomal biogenesis factor 3 | 2745 | 0.054 | 0.5399 | No |
| 14 | SCP2 | SCP2 Entrez, Source | sterol carrier protein 2 | 3534 | 0.042 | 0.4954 | No |
| 15 | GNPAT | GNPAT Entrez, Source | glyceronephosphate O-acyltransferase | 3783 | 0.039 | 0.4905 | No |
| 16 | PEX6 | PEX6 Entrez, Source | peroxisomal biogenesis factor 6 | 3950 | 0.037 | 0.4909 | No |
| 17 | PECI | PECI Entrez, Source | peroxisomal D3,D2-enoyl-CoA isomerase | 4265 | 0.033 | 0.4787 | No |
| 18 | HSD17B4 | HSD17B4 Entrez, Source | hydroxysteroid (17-beta) dehydrogenase 4 | 4370 | 0.032 | 0.4819 | No |
| 19 | PEX11B | PEX11B Entrez, Source | peroxisomal biogenesis factor 11B | 4699 | 0.028 | 0.4668 | No |
| 20 | ACOT8 | ACOT8 Entrez, Source | acyl-CoA thioesterase 8 | 4705 | 0.027 | 0.4760 | No |
| 21 | ACOX3 | ACOX3 Entrez, Source | acyl-Coenzyme A oxidase 3, pristanoyl | 4732 | 0.027 | 0.4834 | No |
| 22 | PHYH | PHYH Entrez, Source | phytanoyl-CoA 2-hydroxylase | 4827 | 0.026 | 0.4854 | No |
| 23 | HAO2 | HAO2 Entrez, Source | hydroxyacid oxidase 2 (long chain) | 4991 | 0.024 | 0.4815 | No |
| 24 | PEX14 | PEX14 Entrez, Source | peroxisomal biogenesis factor 14 | 5019 | 0.024 | 0.4876 | No |
| 25 | PRDX5 | PRDX5 Entrez, Source | peroxiredoxin 5 | 6258 | 0.010 | 0.3981 | No |
| 26 | PEX7 | PEX7 Entrez, Source | peroxisomal biogenesis factor 7 | 7894 | -0.008 | 0.2781 | No |
| 27 | ECH1 | ECH1 Entrez, Source | enoyl Coenzyme A hydratase 1, peroxisomal | 8150 | -0.011 | 0.2628 | No |
| 28 | AGPS | AGPS Entrez, Source | alkylglycerone phosphate synthase | 8182 | -0.012 | 0.2645 | No |
| 29 | ISOC1 | ISOC1 Entrez, Source | isochorismatase domain containing 1 | 8281 | -0.013 | 0.2616 | No |
| 30 | FIS1 | FIS1 Entrez, Source | fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) | 8518 | -0.016 | 0.2494 | No |
| 31 | ABCD2 | ABCD2 Entrez, Source | ATP-binding cassette, sub-family D (ALD), member 2 | 10245 | -0.041 | 0.1337 | No |
| 32 | PXMP3 | PXMP3 Entrez, Source | peroxisomal membrane protein 3, 35kDa (Zellweger syndrome) | 10467 | -0.045 | 0.1326 | No |
| 33 | ABCD3 | ABCD3 Entrez, Source | ATP-binding cassette, sub-family D (ALD), member 3 | 10872 | -0.053 | 0.1207 | No |
| 34 | IDE | IDE Entrez, Source | insulin-degrading enzyme | 10957 | -0.055 | 0.1334 | No |
| 35 | CROT | CROT Entrez, Source | carnitine O-octanoyltransferase | 12676 | -0.132 | 0.0500 | No |