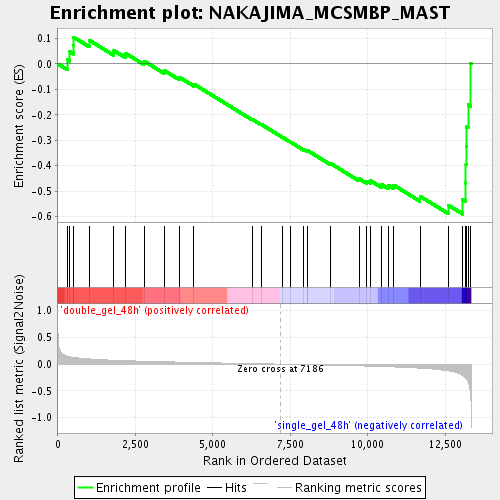
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | single_gel_48h |
| GeneSet | NAKAJIMA_MCSMBP_MAST |
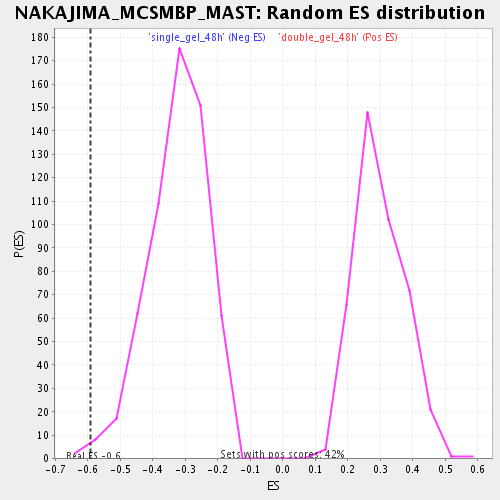
| Enrichment Score (ES) | -0.5906481 |
| Normalized Enrichment Score (NES) | -1.8159405 |
| Nominal p-value | 0.008547009 |
| FDR q-value | 0.04013282 |
| FWER p-Value | 0.948 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ALAS1 | ALAS1 Entrez, Source | aminolevulinate, delta-, synthase 1 | 300 | 0.149 | 0.0174 | No |
| 2 | HDC | HDC Entrez, Source | histidine decarboxylase | 383 | 0.138 | 0.0480 | No |
| 3 | CYP1B1 | CYP1B1 Entrez, Source | cytochrome P450, family 1, subfamily B, polypeptide 1 | 506 | 0.124 | 0.0721 | No |
| 4 | ENPP2 | ENPP2 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) | 511 | 0.124 | 0.1050 | No |
| 5 | GDF15 | GDF15 Entrez, Source | growth differentiation factor 15 | 1016 | 0.097 | 0.0930 | No |
| 6 | TPSAB1 | TPSAB1 Entrez, Source | tryptase alpha/beta 1 | 1806 | 0.072 | 0.0529 | No |
| 7 | TSPAN4 | TSPAN4 Entrez, Source | tetraspanin 4 | 2173 | 0.064 | 0.0426 | No |
| 8 | ENO2 | ENO2 Entrez, Source | enolase 2 (gamma, neuronal) | 2779 | 0.054 | 0.0115 | No |
| 9 | FCER1A | FCER1A Entrez, Source | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide | 3426 | 0.044 | -0.0253 | No |
| 10 | HPGD | HPGD Entrez, Source | hydroxyprostaglandin dehydrogenase 15-(NAD) | 3915 | 0.038 | -0.0519 | No |
| 11 | CALB2 | CALB2 Entrez, Source | calbindin 2, 29kDa (calretinin) | 4390 | 0.031 | -0.0791 | No |
| 12 | CPA3 | CPA3 Entrez, Source | carboxypeptidase A3 (mast cell) | 6266 | 0.010 | -0.2173 | No |
| 13 | MT3 | MT3 Entrez, Source | metallothionein 3 (growth inhibitory factor (neurotrophic)) | 6558 | 0.007 | -0.2373 | No |
| 14 | PEBP1 | PEBP1 Entrez, Source | phosphatidylethanolamine binding protein 1 | 7255 | -0.001 | -0.2894 | No |
| 15 | CMA1 | CMA1 Entrez, Source | chymase 1, mast cell | 7510 | -0.004 | -0.3074 | No |
| 16 | LTC4S | LTC4S Entrez, Source | leukotriene C4 synthase | 7935 | -0.009 | -0.3370 | No |
| 17 | GATA2 | GATA2 Entrez, Source | GATA binding protein 2 | 8055 | -0.010 | -0.3432 | No |
| 18 | PRDX1 | PRDX1 Entrez, Source | peroxiredoxin 1 | 8062 | -0.010 | -0.3409 | No |
| 19 | MAOB | MAOB Entrez, Source | monoamine oxidase B | 8794 | -0.019 | -0.3907 | No |
| 20 | CTSD | CTSD Entrez, Source | cathepsin D (lysosomal aspartyl peptidase) | 9720 | -0.032 | -0.4516 | No |
| 21 | KIT | KIT Entrez, Source | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | 9974 | -0.036 | -0.4609 | No |
| 22 | CAPG | CAPG Entrez, Source | capping protein (actin filament), gelsolin-like | 10094 | -0.038 | -0.4597 | No |
| 23 | HSP90AB1 | HSP90AB1 Entrez, Source | heat shock protein 90kDa alpha (cytosolic), class B member 1 | 10450 | -0.044 | -0.4745 | No |
| 24 | MT1A | MT1A Entrez, Source | metallothionein 1A (functional) | 10667 | -0.048 | -0.4779 | No |
| 25 | MS4A2 | MS4A2 Entrez, Source | membrane-spanning 4-domains, subfamily A, member 2 (Fc fragment of IgE, high affinity I, receptor for; beta polypeptide) | 10839 | -0.052 | -0.4767 | No |
| 26 | CLU | CLU Entrez, Source | clusterin | 11686 | -0.075 | -0.5203 | No |
| 27 | VAT1 | VAT1 Entrez, Source | vesicle amine transport protein 1 homolog (T californica) | 12596 | -0.125 | -0.5552 | Yes |
| 28 | RGS10 | RGS10 Entrez, Source | regulator of G-protein signalling 10 | 13069 | -0.218 | -0.5326 | Yes |
| 29 | LGALS3 | LGALS3 Entrez, Source | lectin, galactoside-binding, soluble, 3 (galectin 3) | 13164 | -0.267 | -0.4682 | Yes |
| 30 | GPNMB | GPNMB Entrez, Source | glycoprotein (transmembrane) nmb | 13168 | -0.269 | -0.3966 | Yes |
| 31 | FST | FST Entrez, Source | follistatin | 13176 | -0.277 | -0.3231 | Yes |
| 32 | TIMP1 | TIMP1 Entrez, Source | TIMP metallopeptidase inhibitor 1 | 13187 | -0.286 | -0.2476 | Yes |
| 33 | PLAT | PLAT Entrez, Source | plasminogen activator, tissue | 13241 | -0.343 | -0.1599 | Yes |
| 34 | CCL2 | CCL2 Entrez, Source | chemokine (C-C motif) ligand 2 | 13326 | -0.627 | 0.0012 | Yes |