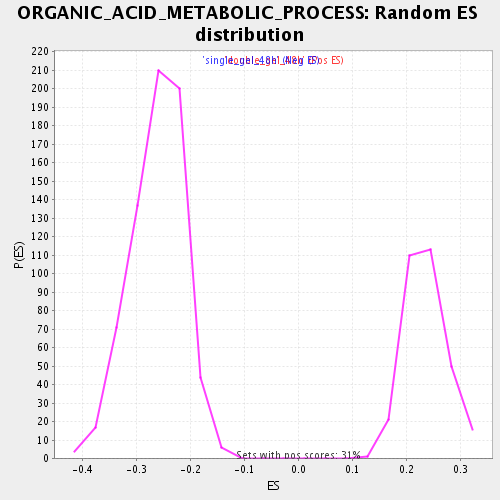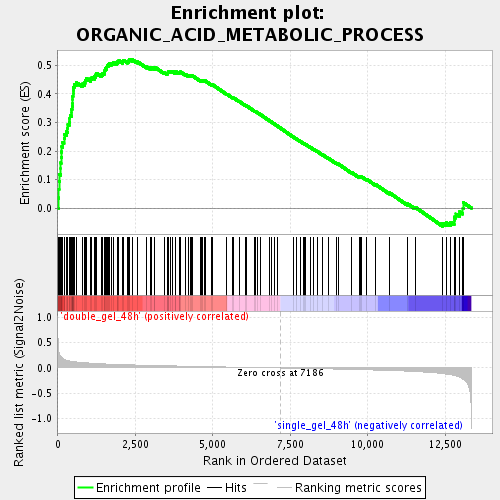
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | ORGANIC_ACID_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.5218259 |
| Normalized Enrichment Score (NES) | 2.2203963 |
| Nominal p-value | 0.0 |
| FDR q-value | 4.5362228E-4 |
| FWER p-Value | 0.0080 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SLC27A5 | SLC27A5 Entrez, Source | solute carrier family 27 (fatty acid transporter), member 5 | 12 | 0.395 | 0.0358 | Yes |
| 2 | FTCD | FTCD Entrez, Source | formiminotransferase cyclodeaminase | 27 | 0.336 | 0.0659 | Yes |
| 3 | BBOX1 | BBOX1 Entrez, Source | butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 | 37 | 0.303 | 0.0933 | Yes |
| 4 | CDO1 | CDO1 Entrez, Source | cysteine dioxygenase, type I | 54 | 0.279 | 0.1180 | Yes |
| 5 | SC4MOL | SC4MOL Entrez, Source | sterol-C4-methyl oxidase-like | 90 | 0.236 | 0.1373 | Yes |
| 6 | HACL1 | HACL1 Entrez, Source | 2-hydroxyacyl-CoA lyase 1 | 94 | 0.229 | 0.1583 | Yes |
| 7 | GLYAT | GLYAT Entrez, Source | glycine-N-acyltransferase | 102 | 0.225 | 0.1786 | Yes |
| 8 | ARG1 | ARG1 Entrez, Source | arginase, liver | 117 | 0.209 | 0.1969 | Yes |
| 9 | ACLY | ACLY Entrez, Source | ATP citrate lyase | 127 | 0.202 | 0.2150 | Yes |
| 10 | PPARA | PPARA Entrez, Source | peroxisome proliferative activated receptor, alpha | 155 | 0.190 | 0.2307 | Yes |
| 11 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 197 | 0.173 | 0.2437 | Yes |
| 12 | GCK | GCK Entrez, Source | glucokinase (hexokinase 4, maturity onset diabetes of the young 2) | 218 | 0.170 | 0.2579 | Yes |
| 13 | ASL | ASL Entrez, Source | argininosuccinate lyase | 273 | 0.154 | 0.2681 | Yes |
| 14 | ALDH6A1 | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 314 | 0.148 | 0.2788 | Yes |
| 15 | MAT1A | MAT1A Entrez, Source | methionine adenosyltransferase I, alpha | 323 | 0.146 | 0.2917 | Yes |
| 16 | FADS2 | FADS2 Entrez, Source | fatty acid desaturase 2 | 362 | 0.140 | 0.3019 | Yes |
| 17 | HDC | HDC Entrez, Source | histidine decarboxylase | 383 | 0.138 | 0.3132 | Yes |
| 18 | NR1H4 | NR1H4 Entrez, Source | nuclear receptor subfamily 1, group H, member 4 | 390 | 0.137 | 0.3254 | Yes |
| 19 | ACOX2 | ACOX2 Entrez, Source | acyl-Coenzyme A oxidase 2, branched chain | 442 | 0.130 | 0.3337 | Yes |
| 20 | TST | TST Entrez, Source | thiosulfate sulfurtransferase (rhodanese) | 446 | 0.130 | 0.3455 | Yes |
| 21 | HGD | HGD Entrez, Source | homogentisate 1,2-dioxygenase (homogentisate oxidase) | 457 | 0.129 | 0.3568 | Yes |
| 22 | CPT1A | CPT1A Entrez, Source | carnitine palmitoyltransferase 1A (liver) | 469 | 0.128 | 0.3678 | Yes |
| 23 | PFKFB1 | PFKFB1 Entrez, Source | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | 476 | 0.127 | 0.3792 | Yes |
| 24 | AS3MT | AS3MT Entrez, Source | arsenic (+3 oxidation state) methyltransferase | 480 | 0.127 | 0.3907 | Yes |
| 25 | FAH | FAH Entrez, Source | fumarylacetoacetate hydrolase (fumarylacetoacetase) | 489 | 0.126 | 0.4018 | Yes |
| 26 | SLC7A2 | SLC7A2 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | 494 | 0.125 | 0.4131 | Yes |
| 27 | ALDH1L1 | ALDH1L1 Entrez, Source | aldehyde dehydrogenase 1 family, member L1 | 502 | 0.125 | 0.4242 | Yes |
| 28 | ACADVL | ACADVL Entrez, Source | acyl-Coenzyme A dehydrogenase, very long chain | 526 | 0.123 | 0.4339 | Yes |
| 29 | AGXT | AGXT Entrez, Source | alanine-glyoxylate aminotransferase (oxalosis I; hyperoxaluria I; glycolicaciduria; serine-pyruvate aminotransferase) | 591 | 0.118 | 0.4400 | Yes |
| 30 | SLC25A10 | SLC25A10 Entrez, Source | solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 | 790 | 0.107 | 0.4350 | Yes |
| 31 | BCKDK | BCKDK Entrez, Source | branched chain ketoacid dehydrogenase kinase | 866 | 0.104 | 0.4390 | Yes |
| 32 | PTS | PTS Entrez, Source | 6-pyruvoyltetrahydropterin synthase | 877 | 0.104 | 0.4478 | Yes |
| 33 | GAMT | GAMT Entrez, Source | guanidinoacetate N-methyltransferase | 910 | 0.102 | 0.4549 | Yes |
| 34 | SCLY | SCLY Entrez, Source | selenocysteine lyase | 1056 | 0.095 | 0.4527 | Yes |
| 35 | CD74 | CD74 Entrez, Source | CD74 molecule, major histocompatibility complex, class II invariant chain | 1097 | 0.093 | 0.4584 | Yes |
| 36 | SLC25A15 | SLC25A15 Entrez, Source | solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 | 1184 | 0.090 | 0.4602 | Yes |
| 37 | GAD2 | GAD2 Entrez, Source | glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) | 1212 | 0.089 | 0.4665 | Yes |
| 38 | GCLC | GCLC Entrez, Source | glutamate-cysteine ligase, catalytic subunit | 1237 | 0.088 | 0.4729 | Yes |
| 39 | MSRA | MSRA Entrez, Source | methionine sulfoxide reductase A | 1404 | 0.082 | 0.4679 | Yes |
| 40 | GNE | GNE Entrez, Source | glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase | 1441 | 0.081 | 0.4728 | Yes |
| 41 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 1493 | 0.080 | 0.4763 | Yes |
| 42 | PAH | PAH Entrez, Source | phenylalanine hydroxylase | 1513 | 0.079 | 0.4822 | Yes |
| 43 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 1528 | 0.079 | 0.4884 | Yes |
| 44 | UGDH | UGDH Entrez, Source | UDP-glucose dehydrogenase | 1562 | 0.078 | 0.4932 | Yes |
| 45 | SLC7A8 | SLC7A8 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 | 1584 | 0.077 | 0.4988 | Yes |
| 46 | BAAT | BAAT Entrez, Source | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 1625 | 0.077 | 0.5028 | Yes |
| 47 | NANP | NANP Entrez, Source | N-acetylneuraminic acid phosphatase | 1668 | 0.075 | 0.5066 | Yes |
| 48 | ACN9 | ACN9 Entrez, Source | ACN9 homolog (S. cerevisiae) | 1743 | 0.073 | 0.5078 | Yes |
| 49 | PTGES | PTGES Entrez, Source | prostaglandin E synthase | 1797 | 0.072 | 0.5105 | Yes |
| 50 | YARS | YARS Entrez, Source | tyrosyl-tRNA synthetase | 1907 | 0.070 | 0.5087 | Yes |
| 51 | FADS1 | FADS1 Entrez, Source | fatty acid desaturase 1 | 1932 | 0.070 | 0.5134 | Yes |
| 52 | QDPR | QDPR Entrez, Source | quinoid dihydropteridine reductase | 1967 | 0.069 | 0.5172 | Yes |
| 53 | AKR1D1 | AKR1D1 Entrez, Source | aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) | 2098 | 0.066 | 0.5135 | Yes |
| 54 | FAAH | FAAH Entrez, Source | fatty acid amide hydrolase | 2116 | 0.066 | 0.5183 | Yes |
| 55 | BCKDHA | BCKDHA Entrez, Source | branched chain keto acid dehydrogenase E1, alpha polypeptide | 2252 | 0.063 | 0.5139 | Yes |
| 56 | CYP4F2 | CYP4F2 Entrez, Source | cytochrome P450, family 4, subfamily F, polypeptide 2 | 2291 | 0.062 | 0.5168 | Yes |
| 57 | MLYCD | MLYCD Entrez, Source | malonyl-CoA decarboxylase | 2302 | 0.062 | 0.5218 | Yes |
| 58 | SLC7A5 | SLC7A5 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 2396 | 0.060 | 0.5204 | No |
| 59 | PPARD | PPARD Entrez, Source | peroxisome proliferative activated receptor, delta | 2558 | 0.058 | 0.5135 | No |
| 60 | ASRGL1 | ASRGL1 Entrez, Source | asparaginase like 1 | 2861 | 0.052 | 0.4955 | No |
| 61 | TNFRSF1A | TNFRSF1A Entrez, Source | tumor necrosis factor receptor superfamily, member 1A | 2994 | 0.050 | 0.4902 | No |
| 62 | PTGS1 | PTGS1 Entrez, Source | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | 3007 | 0.050 | 0.4939 | No |
| 63 | AARS | AARS Entrez, Source | alanyl-tRNA synthetase | 3123 | 0.048 | 0.4897 | No |
| 64 | GCSH | GCSH Entrez, Source | glycine cleavage system protein H (aminomethyl carrier) | 3126 | 0.048 | 0.4940 | No |
| 65 | ACOT2 | ACOT2 Entrez, Source | acyl-CoA thioesterase 2 | 3435 | 0.044 | 0.4748 | No |
| 66 | ACADSB | ACADSB Entrez, Source | acyl-Coenzyme A dehydrogenase, short/branched chain | 3526 | 0.043 | 0.4719 | No |
| 67 | AMT | AMT Entrez, Source | aminomethyltransferase | 3552 | 0.042 | 0.4739 | No |
| 68 | GLS2 | GLS2 Entrez, Source | glutaminase 2 (liver, mitochondrial) | 3555 | 0.042 | 0.4777 | No |
| 69 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 3579 | 0.042 | 0.4798 | No |
| 70 | GBA2 | GBA2 Entrez, Source | glucosidase, beta (bile acid) 2 | 3630 | 0.041 | 0.4799 | No |
| 71 | SDS | SDS Entrez, Source | serine dehydratase | 3684 | 0.041 | 0.4797 | No |
| 72 | GNPAT | GNPAT Entrez, Source | glyceronephosphate O-acyltransferase | 3783 | 0.039 | 0.4759 | No |
| 73 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 3799 | 0.039 | 0.4784 | No |
| 74 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 3912 | 0.038 | 0.4734 | No |
| 75 | HPGD | HPGD Entrez, Source | hydroxyprostaglandin dehydrogenase 15-(NAD) | 3915 | 0.038 | 0.4768 | No |
| 76 | IDH1 | IDH1 Entrez, Source | isocitrate dehydrogenase 1 (NADP+), soluble | 3953 | 0.037 | 0.4774 | No |
| 77 | ALDH5A1 | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 4114 | 0.035 | 0.4686 | No |
| 78 | GATM | GATM Entrez, Source | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 4229 | 0.033 | 0.4630 | No |
| 79 | PECI | PECI Entrez, Source | peroxisomal D3,D2-enoyl-CoA isomerase | 4265 | 0.033 | 0.4634 | No |
| 80 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 4313 | 0.032 | 0.4629 | No |
| 81 | IGF1 | IGF1 Entrez, Source | insulin-like growth factor 1 (somatomedin C) | 4345 | 0.032 | 0.4635 | No |
| 82 | SLC7A7 | SLC7A7 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 | 4600 | 0.029 | 0.4470 | No |
| 83 | ASPA | ASPA Entrez, Source | aspartoacylase (Canavan disease) | 4645 | 0.028 | 0.4462 | No |
| 84 | ACO1 | ACO1 Entrez, Source | aconitase 1, soluble | 4655 | 0.028 | 0.4482 | No |
| 85 | ACOX3 | ACOX3 Entrez, Source | acyl-Coenzyme A oxidase 3, pristanoyl | 4732 | 0.027 | 0.4449 | No |
| 86 | INDO | INDO Entrez, Source | indoleamine-pyrrole 2,3 dioxygenase | 4751 | 0.027 | 0.4461 | No |
| 87 | GSS | GSS Entrez, Source | glutathione synthetase | 4963 | 0.024 | 0.4323 | No |
| 88 | HAO2 | HAO2 Entrez, Source | hydroxyacid oxidase 2 (long chain) | 4991 | 0.024 | 0.4325 | No |
| 89 | GAD1 | GAD1 Entrez, Source | glutamate decarboxylase 1 (brain, 67kDa) | 5455 | 0.019 | 0.3992 | No |
| 90 | GGT1 | GGT1 Entrez, Source | gamma-glutamyltransferase 1 | 5638 | 0.017 | 0.3870 | No |
| 91 | NFS1 | NFS1 Entrez, Source | NFS1 nitrogen fixation 1 (S. cerevisiae) | 5674 | 0.016 | 0.3858 | No |
| 92 | ACADM | ACADM Entrez, Source | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain | 5845 | 0.014 | 0.3743 | No |
| 93 | GCLM | GCLM Entrez, Source | glutamate-cysteine ligase, modifier subunit | 6039 | 0.012 | 0.3609 | No |
| 94 | IDH3B | IDH3B Entrez, Source | isocitrate dehydrogenase 3 (NAD+) beta | 6092 | 0.012 | 0.3580 | No |
| 95 | DARS | DARS Entrez, Source | aspartyl-tRNA synthetase | 6355 | 0.009 | 0.3390 | No |
| 96 | GGTLA1 | GGTLA1 Entrez, Source | gamma-glutamyltransferase-like activity 1 | 6376 | 0.009 | 0.3383 | No |
| 97 | ADI1 | ADI1 Entrez, Source | acireductone dioxygenase 1 | 6429 | 0.008 | 0.3352 | No |
| 98 | PPARGC1A | PPARGC1A Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, alpha | 6549 | 0.007 | 0.3268 | No |
| 99 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 6812 | 0.004 | 0.3074 | No |
| 100 | KARS | KARS Entrez, Source | lysyl-tRNA synthetase | 6885 | 0.003 | 0.3022 | No |
| 101 | PTGDS | PTGDS Entrez, Source | prostaglandin D2 synthase 21kDa (brain) | 6985 | 0.002 | 0.2949 | No |
| 102 | ADIPOR2 | ADIPOR2 Entrez, Source | adiponectin receptor 2 | 7084 | 0.001 | 0.2876 | No |
| 103 | SLC3A1 | SLC3A1 Entrez, Source | 1 | 7608 | -0.005 | 0.2485 | No |
| 104 | ACOT12 | ACOT12 Entrez, Source | acyl-CoA thioesterase 12 | 7714 | -0.006 | 0.2411 | No |
| 105 | MPST | MPST Entrez, Source | mercaptopyruvate sulfurtransferase | 7835 | -0.008 | 0.2327 | No |
| 106 | AGPAT1 | AGPAT1 Entrez, Source | 1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) | 7919 | -0.009 | 0.2272 | No |
| 107 | ACADS | ACADS Entrez, Source | acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain | 7949 | -0.009 | 0.2258 | No |
| 108 | WARS | WARS Entrez, Source | tryptophanyl-tRNA synthetase | 7995 | -0.009 | 0.2233 | No |
| 109 | ECH1 | ECH1 Entrez, Source | enoyl Coenzyme A hydratase 1, peroxisomal | 8150 | -0.011 | 0.2127 | No |
| 110 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 8248 | -0.012 | 0.2065 | No |
| 111 | GLUD1 | GLUD1 Entrez, Source | glutamate dehydrogenase 1 | 8367 | -0.014 | 0.1988 | No |
| 112 | HSD3B7 | HSD3B7 Entrez, Source | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 | 8545 | -0.016 | 0.1869 | No |
| 113 | SCYE1 | SCYE1 Entrez, Source | small inducible cytokine subfamily E, member 1 (endothelial monocyte-activating) | 8723 | -0.018 | 0.1752 | No |
| 114 | ARPP-19 | ARPP-19 Entrez, Source | - | 8985 | -0.022 | 0.1574 | No |
| 115 | CPT1B | CPT1B Entrez, Source | carnitine palmitoyltransferase 1B (muscle) | 9047 | -0.023 | 0.1549 | No |
| 116 | FARS2 | FARS2 Entrez, Source | phenylalanine-tRNA synthetase 2 (mitochondrial) | 9474 | -0.029 | 0.1253 | No |
| 117 | SLC7A9 | SLC7A9 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 | 9724 | -0.032 | 0.1095 | No |
| 118 | MCCC2 | MCCC2 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 2 (beta) | 9753 | -0.033 | 0.1104 | No |
| 119 | CYP7B1 | CYP7B1 Entrez, Source | cytochrome P450, family 7, subfamily B, polypeptide 1 | 9803 | -0.033 | 0.1098 | No |
| 120 | LTA4H | LTA4H Entrez, Source | leukotriene A4 hydrolase | 9958 | -0.036 | 0.1015 | No |
| 121 | ABCD2 | ABCD2 Entrez, Source | ATP-binding cassette, sub-family D (ALD), member 2 | 10245 | -0.041 | 0.0836 | No |
| 122 | ADIPOR1 | ADIPOR1 Entrez, Source | adiponectin receptor 1 | 10701 | -0.049 | 0.0537 | No |
| 123 | MIF | MIF Entrez, Source | macrophage migration inhibitory factor (glycosylation-inhibiting factor) | 11278 | -0.062 | 0.0158 | No |
| 124 | DEGS1 | DEGS1 Entrez, Source | degenerative spermatocyte homolog 1, lipid desaturase (Drosophila) | 11526 | -0.069 | 0.0036 | No |
| 125 | PLOD1 | PLOD1 Entrez, Source | procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | 12413 | -0.112 | -0.0531 | No |
| 126 | LYPLA3 | LYPLA3 Entrez, Source | lysophospholipase 3 (lysosomal phospholipase A2) | 12539 | -0.121 | -0.0514 | No |
| 127 | CROT | CROT Entrez, Source | carnitine O-octanoyltransferase | 12676 | -0.132 | -0.0495 | No |
| 128 | BRCA1 | BRCA1 Entrez, Source | breast cancer 1, early onset | 12787 | -0.146 | -0.0442 | No |
| 129 | SLC6A6 | SLC6A6 Entrez, Source | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | 12793 | -0.147 | -0.0310 | No |
| 130 | DDAH1 | DDAH1 Entrez, Source | dimethylarginine dimethylaminohydrolase 1 | 12847 | -0.155 | -0.0206 | No |
| 131 | DDAH2 | DDAH2 Entrez, Source | dimethylarginine dimethylaminohydrolase 2 | 12970 | -0.181 | -0.0131 | No |
| 132 | SMS | SMS Entrez, Source | spermine synthase | 13073 | -0.219 | -0.0005 | No |
| 133 | BCAT1 | BCAT1 Entrez, Source | branched chain aminotransferase 1, cytosolic | 13084 | -0.224 | 0.0195 | No |