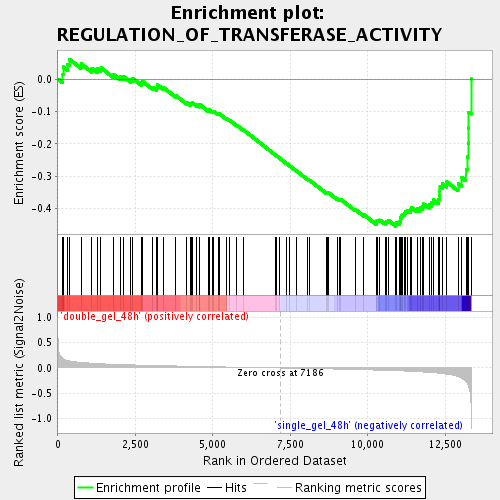
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | single_gel_48h |
| GeneSet | REGULATION_OF_TRANSFERASE_ACTIVITY |
| Enrichment Score (ES) | -0.45477772 |
| Normalized Enrichment Score (NES) | -1.6839134 |
| Nominal p-value | 0.0030721966 |
| FDR q-value | 0.07396859 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 160 | 0.189 | 0.0134 | No |
| 2 | CHRM1 | CHRM1 Entrez, Source | cholinergic receptor, muscarinic 1 | 169 | 0.184 | 0.0376 | No |
| 3 | EGF | EGF Entrez, Source | epidermal growth factor (beta-urogastrone) | 316 | 0.148 | 0.0465 | No |
| 4 | TRIB3 | TRIB3 Entrez, Source | tribbles homolog 3 (Drosophila) | 367 | 0.139 | 0.0615 | No |
| 5 | THY1 | THY1 Entrez, Source | Thy-1 cell surface antigen | 748 | 0.109 | 0.0474 | No |
| 6 | CD4 | CD4 Entrez, Source | CD4 molecule | 1093 | 0.094 | 0.0341 | No |
| 7 | ADRA2A | ADRA2A Entrez, Source | adrenergic, alpha-2A-, receptor | 1280 | 0.087 | 0.0317 | No |
| 8 | CAMKK2 | CAMKK2 Entrez, Source | calcium/calmodulin-dependent protein kinase kinase 2, beta | 1389 | 0.083 | 0.0347 | No |
| 9 | NF1 | NF1 Entrez, Source | neurofibromin 1 (neurofibromatosis, von Recklinghausen disease, Watson disease) | 1803 | 0.072 | 0.0132 | No |
| 10 | GNG3 | GNG3 Entrez, Source | guanine nucleotide binding protein (G protein), gamma 3 | 2009 | 0.068 | 0.0069 | No |
| 11 | GTPBP4 | GTPBP4 Entrez, Source | GTP binding protein 4 | 2114 | 0.066 | 0.0079 | No |
| 12 | C5AR1 | C5AR1 Entrez, Source | complement component 5a receptor 1 | 2348 | 0.061 | -0.0015 | No |
| 13 | SHC1 | SHC1 Entrez, Source | SHC (Src homology 2 domain containing) transforming protein 1 | 2417 | 0.060 | 0.0015 | No |
| 14 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 2688 | 0.055 | -0.0115 | No |
| 15 | RGS3 | RGS3 Entrez, Source | regulator of G-protein signalling 3 | 2720 | 0.055 | -0.0064 | No |
| 16 | PRLR | PRLR Entrez, Source | prolactin receptor | 3062 | 0.049 | -0.0256 | No |
| 17 | CDK5RAP3 | CDK5RAP3 Entrez, Source | CDK5 regulatory subunit associated protein 3 | 3193 | 0.047 | -0.0290 | No |
| 18 | GRM4 | GRM4 Entrez, Source | glutamate receptor, metabotropic 4 | 3208 | 0.047 | -0.0237 | No |
| 19 | HIPK3 | HIPK3 Entrez, Source | homeodomain interacting protein kinase 3 | 3217 | 0.047 | -0.0180 | No |
| 20 | RBL2 | RBL2 Entrez, Source | retinoblastoma-like 2 (p130) | 3391 | 0.045 | -0.0251 | No |
| 21 | GAP43 | GAP43 Entrez, Source | growth associated protein 43 | 3808 | 0.039 | -0.0512 | No |
| 22 | EREG | EREG Entrez, Source | epiregulin | 4157 | 0.034 | -0.0728 | No |
| 23 | PICK1 | PICK1 Entrez, Source | protein interacting with PRKCA 1 | 4289 | 0.033 | -0.0783 | No |
| 24 | CDKN3 | CDKN3 Entrez, Source | cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | 4303 | 0.033 | -0.0749 | No |
| 25 | CBLC | CBLC Entrez, Source | Cas-Br-M (murine) ecotropic retroviral transforming sequence c | 4341 | 0.032 | -0.0734 | No |
| 26 | TGFA | TGFA Entrez, Source | transforming growth factor, alpha | 4483 | 0.030 | -0.0800 | No |
| 27 | CHRNA7 | CHRNA7 Entrez, Source | cholinergic receptor, nicotinic, alpha 7 | 4570 | 0.029 | -0.0825 | No |
| 28 | PTEN | PTEN Entrez, Source | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 4572 | 0.029 | -0.0787 | No |
| 29 | CDC25A | CDC25A Entrez, Source | cell division cycle 25A | 4860 | 0.026 | -0.0969 | No |
| 30 | PROK2 | PROK2 Entrez, Source | prokineticin 2 | 4883 | 0.025 | -0.0952 | No |
| 31 | MAP3K10 | MAP3K10 Entrez, Source | mitogen-activated protein kinase kinase kinase 10 | 4975 | 0.024 | -0.0988 | No |
| 32 | CDKN1C | CDKN1C Entrez, Source | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 5020 | 0.024 | -0.0989 | No |
| 33 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 5178 | 0.022 | -0.1079 | No |
| 34 | CDK5RAP1 | CDK5RAP1 Entrez, Source | CDK5 regulatory subunit associated protein 1 | 5204 | 0.021 | -0.1069 | No |
| 35 | TRIB1 | TRIB1 Entrez, Source | tribbles homolog 1 (Drosophila) | 5445 | 0.019 | -0.1224 | No |
| 36 | ALS2 | ALS2 Entrez, Source | amyotrophic lateral sclerosis 2 (juvenile) | 5536 | 0.018 | -0.1268 | No |
| 37 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 5771 | 0.015 | -0.1424 | No |
| 38 | PARD3 | PARD3 Entrez, Source | par-3 partitioning defective 3 homolog (C. elegans) | 5990 | 0.013 | -0.1572 | No |
| 39 | APC | APC Entrez, Source | adenomatosis polyposis coli | 7029 | 0.002 | -0.2353 | No |
| 40 | TAOK2 | TAOK2 Entrez, Source | TAO kinase 2 | 7066 | 0.001 | -0.2378 | No |
| 41 | CDKN2C | CDKN2C Entrez, Source | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | 7160 | 0.000 | -0.2448 | No |
| 42 | CARTPT | CARTPT Entrez, Source | CART prepropeptide | 7384 | -0.002 | -0.2613 | No |
| 43 | ADRA2C | ADRA2C Entrez, Source | adrenergic, alpha-2C-, receptor | 7484 | -0.004 | -0.2683 | No |
| 44 | DUSP2 | DUSP2 Entrez, Source | dual specificity phosphatase 2 | 7690 | -0.006 | -0.2831 | No |
| 45 | PIK3CB | PIK3CB Entrez, Source | phosphoinositide-3-kinase, catalytic, beta polypeptide | 8066 | -0.010 | -0.3100 | No |
| 46 | EDN2 | EDN2 Entrez, Source | endothelin 2 | 8122 | -0.011 | -0.3127 | No |
| 47 | SERINC1 | SERINC1 Entrez, Source | serine incorporator 1 | 8655 | -0.018 | -0.3505 | No |
| 48 | DAXX | DAXX Entrez, Source | death-associated protein 6 | 8710 | -0.018 | -0.3521 | No |
| 49 | RGS4 | RGS4 Entrez, Source | regulator of G-protein signalling 4 | 8726 | -0.018 | -0.3508 | No |
| 50 | CDK7 | CDK7 Entrez, Source | cyclin-dependent kinase 7 (MO15 homolog, Xenopus laevis, cdk-activating kinase) | 9024 | -0.022 | -0.3702 | No |
| 51 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 9099 | -0.023 | -0.3727 | No |
| 52 | CDK5R1 | CDK5R1 Entrez, Source | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | 9127 | -0.023 | -0.3716 | No |
| 53 | GPS1 | GPS1 Entrez, Source | G protein pathway suppressor 1 | 9588 | -0.030 | -0.4022 | No |
| 54 | LYK5 | LYK5 Entrez, Source | - | 9877 | -0.034 | -0.4193 | No |
| 55 | PKN1 | PKN1 Entrez, Source | protein kinase N1 | 10278 | -0.041 | -0.4440 | No |
| 56 | CCNG1 | CCNG1 Entrez, Source | cyclin G1 | 10280 | -0.041 | -0.4386 | No |
| 57 | SPRED1 | SPRED1 Entrez, Source | sprouty-related, EVH1 domain containing 1 | 10322 | -0.042 | -0.4360 | No |
| 58 | GHRL | GHRL Entrez, Source | ghrelin/obestatin preprohormone | 10393 | -0.043 | -0.4355 | No |
| 59 | CXCR4 | CXCR4 Entrez, Source | chemokine (C-X-C motif) receptor 4 | 10564 | -0.046 | -0.4421 | No |
| 60 | GADD45B | GADD45B Entrez, Source | growth arrest and DNA-damage-inducible, beta | 10614 | -0.047 | -0.4394 | No |
| 61 | MAP3K11 | MAP3K11 Entrez, Source | mitogen-activated protein kinase kinase kinase 11 | 10674 | -0.049 | -0.4373 | No |
| 62 | ERBB2 | ERBB2 Entrez, Source | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | 10906 | -0.054 | -0.4475 | Yes |
| 63 | SPRED2 | SPRED2 Entrez, Source | sprouty-related, EVH1 domain containing 2 | 10933 | -0.054 | -0.4422 | Yes |
| 64 | CDKN2B | CDKN2B Entrez, Source | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 11022 | -0.056 | -0.4412 | Yes |
| 65 | MNAT1 | MNAT1 Entrez, Source | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 11046 | -0.057 | -0.4353 | Yes |
| 66 | PAK1 | PAK1 Entrez, Source | p21/Cdc42/Rac1-activated kinase 1 (STE20 homolog, yeast) | 11055 | -0.057 | -0.4282 | Yes |
| 67 | PPAP2A | PPAP2A Entrez, Source | phosphatidic acid phosphatase type 2A | 11084 | -0.058 | -0.4226 | Yes |
| 68 | DBNL | DBNL Entrez, Source | drebrin-like | 11132 | -0.059 | -0.4182 | Yes |
| 69 | MAP2K3 | MAP2K3 Entrez, Source | mitogen-activated protein kinase kinase 3 | 11186 | -0.060 | -0.4142 | Yes |
| 70 | ADRB2 | ADRB2 Entrez, Source | adrenergic, beta-2-, receptor, surface | 11210 | -0.060 | -0.4078 | Yes |
| 71 | TARBP2 | TARBP2 Entrez, Source | Tar (HIV-1) RNA binding protein 2 | 11281 | -0.062 | -0.4047 | Yes |
| 72 | MADD | MADD Entrez, Source | MAP-kinase activating death domain | 11375 | -0.064 | -0.4031 | Yes |
| 73 | RB1 | RB1 Entrez, Source | retinoblastoma 1 (including osteosarcoma) | 11401 | -0.065 | -0.3962 | Yes |
| 74 | CD81 | CD81 Entrez, Source | CD81 molecule | 11591 | -0.072 | -0.4008 | Yes |
| 75 | FGF2 | FGF2 Entrez, Source | fibroblast growth factor 2 (basic) | 11687 | -0.075 | -0.3979 | Yes |
| 76 | CCND3 | CCND3 Entrez, Source | cyclin D3 | 11760 | -0.077 | -0.3929 | Yes |
| 77 | CDKN2D | CDKN2D Entrez, Source | cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | 11809 | -0.080 | -0.3858 | Yes |
| 78 | CDC37 | CDC37 Entrez, Source | CDC37 cell division cycle 37 homolog (S. cerevisiae) | 11983 | -0.086 | -0.3873 | Yes |
| 79 | CDKN2A | CDKN2A Entrez, Source | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 12064 | -0.090 | -0.3812 | Yes |
| 80 | PAK2 | PAK2 Entrez, Source | p21 (CDKN1A)-activated kinase 2 | 12118 | -0.092 | -0.3728 | Yes |
| 81 | PDCD4 | PDCD4 Entrez, Source | programmed cell death 4 (neoplastic transformation inhibitor) | 12282 | -0.102 | -0.3714 | Yes |
| 82 | ANAPC2 | ANAPC2 Entrez, Source | anaphase promoting complex subunit 2 | 12308 | -0.104 | -0.3593 | Yes |
| 83 | PTPRC | PTPRC Entrez, Source | protein tyrosine phosphatase, receptor type, C | 12324 | -0.105 | -0.3463 | Yes |
| 84 | DUSP6 | DUSP6 Entrez, Source | dual specificity phosphatase 6 | 12330 | -0.106 | -0.3324 | Yes |
| 85 | SERTAD1 | SERTAD1 Entrez, Source | SERTA domain containing 1 | 12410 | -0.112 | -0.3234 | Yes |
| 86 | PLCE1 | PLCE1 Entrez, Source | phospholipase C, epsilon 1 | 12550 | -0.121 | -0.3175 | Yes |
| 87 | CHEK1 | CHEK1 Entrez, Source | CHK1 checkpoint homolog (S. pombe) | 12915 | -0.168 | -0.3223 | Yes |
| 88 | EDG5 | EDG5 Entrez, Source | endothelial differentiation, sphingolipid G-protein-coupled receptor, 5 | 13028 | -0.198 | -0.3041 | Yes |
| 89 | DUSP9 | DUSP9 Entrez, Source | dual specificity phosphatase 9 | 13170 | -0.271 | -0.2783 | Yes |
| 90 | SERINC5 | SERINC5 Entrez, Source | serine incorporator 5 | 13209 | -0.303 | -0.2404 | Yes |
| 91 | ADORA2B | ADORA2B Entrez, Source | adenosine A2b receptor | 13236 | -0.333 | -0.1975 | Yes |
| 92 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 13250 | -0.352 | -0.1511 | Yes |
| 93 | SERINC2 | SERINC2 Entrez, Source | serine incorporator 2 | 13258 | -0.367 | -0.1023 | Yes |
| 94 | CD24 | CD24 Entrez, Source | CD24 molecule | 13336 | -0.806 | 0.0005 | Yes |