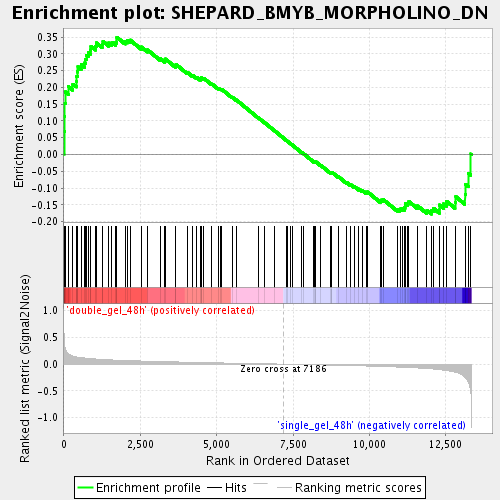
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | SHEPARD_BMYB_MORPHOLINO_DN |
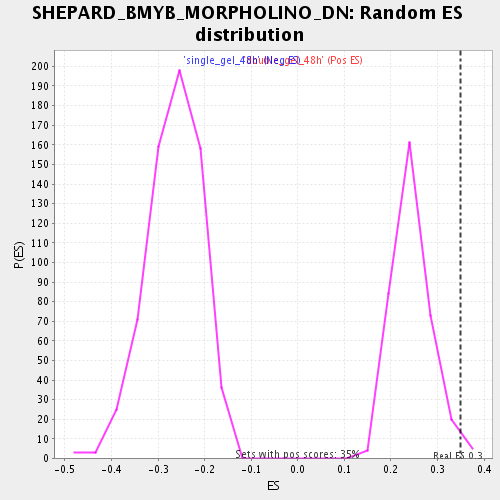
| Enrichment Score (ES) | 0.3497019 |
| Normalized Enrichment Score (NES) | 1.4348034 |
| Nominal p-value | 0.014409222 |
| FDR q-value | 0.18743233 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EDG1 | EDG1 Entrez, Source | endothelial differentiation, sphingolipid G-protein-coupled receptor, 1 | 3 | 0.566 | 0.0683 | Yes |
| 2 | ACAT2 | ACAT2 Entrez, Source | acetyl-Coenzyme A acetyltransferase 2 (acetoacetyl Coenzyme A thiolase) | 14 | 0.377 | 0.1132 | Yes |
| 3 | TM7SF2 | TM7SF2 Entrez, Source | transmembrane 7 superfamily member 2 | 24 | 0.340 | 0.1536 | Yes |
| 4 | CEBPA | CEBPA Entrez, Source | CCAAT/enhancer binding protein (C/EBP), alpha | 46 | 0.289 | 0.1870 | Yes |
| 5 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 148 | 0.193 | 0.2027 | Yes |
| 6 | POU3F1 | POU3F1 Entrez, Source | POU domain, class 3, transcription factor 1 | 291 | 0.150 | 0.2101 | Yes |
| 7 | RBP4 | RBP4 Entrez, Source | retinol binding protein 4, plasma | 409 | 0.134 | 0.2176 | Yes |
| 8 | VTN | VTN Entrez, Source | vitronectin | 420 | 0.133 | 0.2329 | Yes |
| 9 | GCH1 | GCH1 Entrez, Source | GTP cyclohydrolase 1 (dopa-responsive dystonia) | 441 | 0.131 | 0.2472 | Yes |
| 10 | IGFBP1 | IGFBP1 Entrez, Source | insulin-like growth factor binding protein 1 | 459 | 0.129 | 0.2615 | Yes |
| 11 | SARDH | SARDH Entrez, Source | sarcosine dehydrogenase | 566 | 0.121 | 0.2681 | Yes |
| 12 | MDM4 | MDM4 Entrez, Source | Mdm4, transformed 3T3 cell double minute 4, p53 binding protein (mouse) | 683 | 0.113 | 0.2730 | Yes |
| 13 | PRODH2 | PRODH2 Entrez, Source | proline dehydrogenase (oxidase) 2 | 708 | 0.111 | 0.2846 | Yes |
| 14 | PVALB | PVALB Entrez, Source | parvalbumin | 738 | 0.109 | 0.2957 | Yes |
| 15 | SLC6A13 | SLC6A13 Entrez, Source | solute carrier family 6 (neurotransmitter transporter, GABA), member 13 | 797 | 0.107 | 0.3043 | Yes |
| 16 | SERPINC1 | SERPINC1 Entrez, Source | serpin peptidase inhibitor, clade C (antithrombin), member 1 | 864 | 0.104 | 0.3119 | Yes |
| 17 | DHFR | DHFR Entrez, Source | dihydrofolate reductase | 883 | 0.103 | 0.3231 | Yes |
| 18 | POU3F2 | POU3F2 Entrez, Source | POU domain, class 3, transcription factor 2 | 1039 | 0.096 | 0.3229 | Yes |
| 19 | SLC2A5 | SLC2A5 Entrez, Source | solute carrier family 2 (facilitated glucose/fructose transporter), member 5 | 1059 | 0.095 | 0.3330 | Yes |
| 20 | FGA | FGA Entrez, Source | fibrinogen alpha chain | 1253 | 0.088 | 0.3290 | Yes |
| 21 | SLC29A2 | SLC29A2 Entrez, Source | solute carrier family 29 (nucleoside transporters), member 2 | 1277 | 0.087 | 0.3378 | Yes |
| 22 | FGFR4 | FGFR4 Entrez, Source | fibroblast growth factor receptor 4 | 1473 | 0.080 | 0.3328 | Yes |
| 23 | FGG | FGG Entrez, Source | fibrinogen gamma chain | 1572 | 0.078 | 0.3348 | Yes |
| 24 | TTN | TTN Entrez, Source | titin | 1695 | 0.074 | 0.3345 | Yes |
| 25 | C3 | C3 Entrez, Source | complement component 3 | 1731 | 0.074 | 0.3408 | Yes |
| 26 | NR0B2 | NR0B2 Entrez, Source | nuclear receptor subfamily 0, group B, member 2 | 1732 | 0.074 | 0.3497 | Yes |
| 27 | COL1A2 | COL1A2 Entrez, Source | collagen, type I, alpha 2 | 2001 | 0.068 | 0.3377 | No |
| 28 | RTKN | RTKN Entrez, Source | rhotekin | 2071 | 0.066 | 0.3405 | No |
| 29 | BMS1L | BMS1L Entrez, Source | BMS1-like, ribosome assembly protein (yeast) | 2162 | 0.065 | 0.3416 | No |
| 30 | LHX1 | LHX1 Entrez, Source | LIM homeobox 1 | 2531 | 0.058 | 0.3208 | No |
| 31 | GPSM1 | GPSM1 Entrez, Source | G-protein signalling modulator 1 (AGS3-like, C. elegans) | 2735 | 0.055 | 0.3121 | No |
| 32 | DLX1 | DLX1 Entrez, Source | distal-less homeobox 1 | 3150 | 0.048 | 0.2866 | No |
| 33 | IGFBP2 | IGFBP2 Entrez, Source | insulin-like growth factor binding protein 2, 36kDa | 3303 | 0.046 | 0.2807 | No |
| 34 | GRIK3 | GRIK3 Entrez, Source | glutamate receptor, ionotropic, kainate 3 | 3319 | 0.046 | 0.2850 | No |
| 35 | NOTCH2 | NOTCH2 Entrez, Source | Notch homolog 2 (Drosophila) | 3652 | 0.041 | 0.2649 | No |
| 36 | MDK | MDK Entrez, Source | midkine (neurite growth-promoting factor 2) | 3666 | 0.041 | 0.2689 | No |
| 37 | AQP1 | AQP1 Entrez, Source | aquaporin 1 (Colton blood group) | 4037 | 0.036 | 0.2453 | No |
| 38 | CEBPD | CEBPD Entrez, Source | CCAAT/enhancer binding protein (C/EBP), delta | 4223 | 0.034 | 0.2354 | No |
| 39 | OTP | OTP Entrez, Source | orthopedia homolog (Drosophila) | 4337 | 0.032 | 0.2307 | No |
| 40 | COL1A1 | COL1A1 Entrez, Source | collagen, type I, alpha 1 | 4462 | 0.030 | 0.2251 | No |
| 41 | NR2F1 | NR2F1 Entrez, Source | nuclear receptor subfamily 2, group F, member 1 | 4499 | 0.030 | 0.2260 | No |
| 42 | PSAT1 | PSAT1 Entrez, Source | phosphoserine aminotransferase 1 | 4505 | 0.030 | 0.2292 | No |
| 43 | PRKCB1 | PRKCB1 Entrez, Source | protein kinase C, beta 1 | 4579 | 0.029 | 0.2272 | No |
| 44 | SCIN | SCIN Entrez, Source | scinderin | 4841 | 0.026 | 0.2106 | No |
| 45 | PFKFB4 | PFKFB4 Entrez, Source | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 | 5052 | 0.023 | 0.1976 | No |
| 46 | SULF1 | SULF1 Entrez, Source | sulfatase 1 | 5121 | 0.022 | 0.1952 | No |
| 47 | PYGL | PYGL Entrez, Source | phosphorylase, glycogen; liver (Hers disease, glycogen storage disease type VI) | 5160 | 0.022 | 0.1949 | No |
| 48 | HEY1 | HEY1 Entrez, Source | hairy/enhancer-of-split related with YRPW motif 1 | 5501 | 0.018 | 0.1715 | No |
| 49 | TOP1MT | TOP1MT Entrez, Source | topoisomerase (DNA) I, mitochondrial | 5642 | 0.017 | 0.1629 | No |
| 50 | ATP1A1 | ATP1A1 Entrez, Source | ATPase, Na+/K+ transporting, alpha 1 polypeptide | 6356 | 0.009 | 0.1102 | No |
| 51 | PMP22 | PMP22 Entrez, Source | peripheral myelin protein 22 | 6574 | 0.007 | 0.0946 | No |
| 52 | CDC20 | CDC20 Entrez, Source | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 6894 | 0.003 | 0.0709 | No |
| 53 | NUMA1 | NUMA1 Entrez, Source | nuclear mitotic apparatus protein 1 | 7275 | -0.001 | 0.0423 | No |
| 54 | SOX11 | SOX11 Entrez, Source | SRY (sex determining region Y)-box 11 | 7315 | -0.002 | 0.0396 | No |
| 55 | APOE | APOE Entrez, Source | apolipoprotein E | 7411 | -0.003 | 0.0327 | No |
| 56 | CDK2 | CDK2 Entrez, Source | cyclin-dependent kinase 2 | 7469 | -0.003 | 0.0288 | No |
| 57 | CENTB2 | CENTB2 Entrez, Source | centaurin, beta 2 | 7774 | -0.007 | 0.0067 | No |
| 58 | SFRP1 | SFRP1 Entrez, Source | secreted frizzled-related protein 1 | 7830 | -0.007 | 0.0034 | No |
| 59 | SYNGR1 | SYNGR1 Entrez, Source | synaptogyrin 1 | 8181 | -0.012 | -0.0216 | No |
| 60 | CTSL | CTSL Entrez, Source | cathepsin L | 8194 | -0.012 | -0.0211 | No |
| 61 | ORC4L | ORC4L Entrez, Source | origin recognition complex, subunit 4-like (yeast) | 8232 | -0.012 | -0.0224 | No |
| 62 | TXNIP | TXNIP Entrez, Source | thioredoxin interacting protein | 8238 | -0.012 | -0.0213 | No |
| 63 | RAX | RAX Entrez, Source | retina and anterior neural fold homeobox | 8408 | -0.014 | -0.0323 | No |
| 64 | RGS4 | RGS4 Entrez, Source | regulator of G-protein signalling 4 | 8726 | -0.018 | -0.0540 | No |
| 65 | APOA4 | APOA4 Entrez, Source | apolipoprotein A-IV | 8754 | -0.019 | -0.0538 | No |
| 66 | DNMT1 | DNMT1 Entrez, Source | DNA (cytosine-5-)-methyltransferase 1 | 8763 | -0.019 | -0.0521 | No |
| 67 | MYH8 | MYH8 Entrez, Source | myosin, heavy chain 8, skeletal muscle, perinatal | 8972 | -0.022 | -0.0652 | No |
| 68 | FOXM1 | FOXM1 Entrez, Source | forkhead box M1 | 9257 | -0.025 | -0.0836 | No |
| 69 | LAMP2 | LAMP2 Entrez, Source | lysosomal-associated membrane protein 2 | 9376 | -0.027 | -0.0893 | No |
| 70 | PDCL | PDCL Entrez, Source | phosducin-like | 9499 | -0.029 | -0.0950 | No |
| 71 | AQP3 | AQP3 Entrez, Source | aquaporin 3 (Gill blood group) | 9650 | -0.031 | -0.1025 | No |
| 72 | UCP2 | UCP2 Entrez, Source | uncoupling protein 2 (mitochondrial, proton carrier) | 9763 | -0.033 | -0.1070 | No |
| 73 | DUT | DUT Entrez, Source | dUTP pyrophosphatase | 9896 | -0.035 | -0.1128 | No |
| 74 | ITGB4 | ITGB4 Entrez, Source | integrin, beta 4 | 9935 | -0.035 | -0.1113 | No |
| 75 | DCK | DCK Entrez, Source | deoxycytidine kinase | 10358 | -0.043 | -0.1381 | No |
| 76 | SLC4A2 | SLC4A2 Entrez, Source | solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1) | 10395 | -0.043 | -0.1355 | No |
| 77 | POU3F3 | POU3F3 Entrez, Source | POU domain, class 3, transcription factor 3 | 10454 | -0.044 | -0.1345 | No |
| 78 | DDX17 | DDX17 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | 10913 | -0.054 | -0.1626 | No |
| 79 | CDCA2 | CDCA2 Entrez, Source | cell division cycle associated 2 | 10999 | -0.056 | -0.1622 | No |
| 80 | NDE1 | NDE1 Entrez, Source | nudE nuclear distribution gene E homolog 1 (A. nidulans) | 11073 | -0.057 | -0.1608 | No |
| 81 | GOLGB1 | GOLGB1 Entrez, Source | golgi autoantigen, golgin subfamily b, macrogolgin (with transmembrane signal), 1 | 11143 | -0.059 | -0.1589 | No |
| 82 | INPP4A | INPP4A Entrez, Source | inositol polyphosphate-4-phosphatase, type I, 107kDa | 11164 | -0.059 | -0.1532 | No |
| 83 | TCN2 | TCN2 Entrez, Source | transcobalamin II; macrocytic anemia | 11172 | -0.060 | -0.1465 | No |
| 84 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 11253 | -0.061 | -0.1451 | No |
| 85 | PTPRN | PTPRN Entrez, Source | protein tyrosine phosphatase, receptor type, N | 11289 | -0.062 | -0.1403 | No |
| 86 | HMGA1 | HMGA1 Entrez, Source | high mobility group AT-hook 1 | 11574 | -0.071 | -0.1531 | No |
| 87 | DAP | DAP Entrez, Source | death-associated protein | 11879 | -0.082 | -0.1661 | No |
| 88 | CLDN1 | CLDN1 Entrez, Source | claudin 1 | 12033 | -0.089 | -0.1669 | No |
| 89 | PRIM1 | PRIM1 Entrez, Source | primase, polypeptide 1, 49kDa | 12091 | -0.091 | -0.1602 | No |
| 90 | MSH2 | MSH2 Entrez, Source | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | 12287 | -0.103 | -0.1625 | No |
| 91 | SFRS9 | SFRS9 Entrez, Source | splicing factor, arginine/serine-rich 9 | 12292 | -0.103 | -0.1503 | No |
| 92 | ASCL1 | ASCL1 Entrez, Source | achaete-scute complex-like 1 (Drosophila) | 12434 | -0.113 | -0.1473 | No |
| 93 | LIG1 | LIG1 Entrez, Source | ligase I, DNA, ATP-dependent | 12525 | -0.119 | -0.1396 | No |
| 94 | TGM1 | TGM1 Entrez, Source | transglutaminase 1 (K polypeptide epidermal type I, protein-glutamine-gamma-glutamyltransferase) | 12800 | -0.148 | -0.1425 | No |
| 95 | TACC3 | TACC3 Entrez, Source | transforming, acidic coiled-coil containing protein 3 | 12826 | -0.152 | -0.1260 | No |
| 96 | CCNB1 | CCNB1 Entrez, Source | cyclin B1 | 13126 | -0.245 | -0.1189 | No |
| 97 | FABP7 | FABP7 Entrez, Source | fatty acid binding protein 7, brain | 13148 | -0.256 | -0.0895 | No |
| 98 | AURKB | AURKB Entrez, Source | aurora kinase B | 13237 | -0.335 | -0.0556 | No |
| 99 | RGS2 | RGS2 Entrez, Source | regulator of G-protein signalling 2, 24kDa | 13320 | -0.525 | 0.0017 | No |