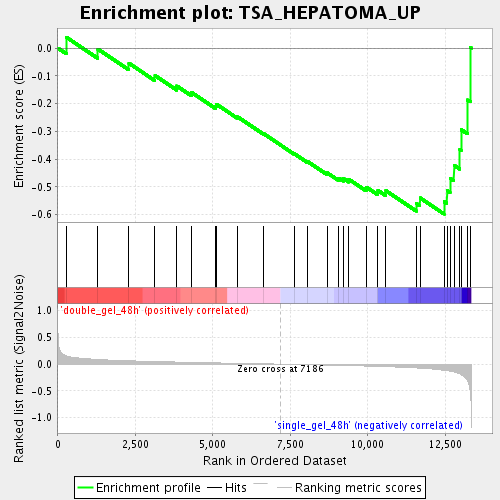
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | single_gel_48h |
| GeneSet | TSA_HEPATOMA_UP |
| Enrichment Score (ES) | -0.5979351 |
| Normalized Enrichment Score (NES) | -1.7470633 |
| Nominal p-value | 0.0034965035 |
| FDR q-value | 0.05039765 |
| FWER p-Value | 0.999 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | H1F0 | H1F0 Entrez, Source | H1 histone family, member 0 | 274 | 0.154 | 0.0393 | No |
| 2 | NIPSNAP1 | NIPSNAP1 Entrez, Source | nipsnap homolog 1 (C. elegans) | 1288 | 0.086 | -0.0032 | No |
| 3 | CYP4F2 | CYP4F2 Entrez, Source | cytochrome P450, family 4, subfamily F, polypeptide 2 | 2291 | 0.062 | -0.0542 | No |
| 4 | SDC4 | SDC4 Entrez, Source | syndecan 4 (amphiglycan, ryudocan) | 3132 | 0.048 | -0.0985 | No |
| 5 | H3F3B | H3F3B Entrez, Source | H3 histone, family 3B (H3.3B) | 3836 | 0.039 | -0.1362 | No |
| 6 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 4313 | 0.032 | -0.1593 | No |
| 7 | ZDHHC9 | ZDHHC9 Entrez, Source | zinc finger, DHHC-type containing 9 | 5099 | 0.023 | -0.2095 | No |
| 8 | DUSP1 | DUSP1 Entrez, Source | dual specificity phosphatase 1 | 5117 | 0.022 | -0.2020 | No |
| 9 | CDKN1A | CDKN1A Entrez, Source | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 5802 | 0.015 | -0.2476 | No |
| 10 | BSCL2 | BSCL2 Entrez, Source | Bernardinelli-Seip congenital lipodystrophy 2 (seipin) | 6646 | 0.006 | -0.3086 | No |
| 11 | ZBED3 | ZBED3 Entrez, Source | zinc finger, BED-type containing 3 | 7625 | -0.005 | -0.3801 | No |
| 12 | PRDX1 | PRDX1 Entrez, Source | peroxiredoxin 1 | 8062 | -0.010 | -0.4089 | No |
| 13 | OPTN | OPTN Entrez, Source | optineurin | 8689 | -0.018 | -0.4490 | No |
| 14 | STAT1 | STAT1 Entrez, Source | signal transducer and activator of transcription 1, 91kDa | 9067 | -0.023 | -0.4684 | No |
| 15 | BSG | BSG Entrez, Source | basigin (Ok blood group) | 9210 | -0.025 | -0.4696 | No |
| 16 | ID2 | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 9384 | -0.027 | -0.4720 | No |
| 17 | LYPLA2 | LYPLA2 Entrez, Source | lysophospholipase II | 9951 | -0.036 | -0.5006 | No |
| 18 | IFITM3 | IFITM3 Entrez, Source | interferon induced transmembrane protein 3 (1-8U) | 10326 | -0.042 | -0.5123 | No |
| 19 | RHOA | RHOA Entrez, Source | ras homolog gene family, member A | 10584 | -0.047 | -0.5135 | No |
| 20 | ATP6AP2 | ATP6AP2 Entrez, Source | ATPase, H+ transporting, lysosomal accessory protein 2 | 11566 | -0.071 | -0.5597 | Yes |
| 21 | CLU | CLU Entrez, Source | clusterin | 11686 | -0.075 | -0.5395 | Yes |
| 22 | JMJD1C | JMJD1C Entrez, Source | jumonji domain containing 1C | 12465 | -0.115 | -0.5531 | Yes |
| 23 | CYR61 | CYR61 Entrez, Source | cysteine-rich, angiogenic inducer, 61 | 12557 | -0.122 | -0.5125 | Yes |
| 24 | TPP1 | TPP1 Entrez, Source | tripeptidyl peptidase I | 12658 | -0.131 | -0.4692 | Yes |
| 25 | TGM2 | TGM2 Entrez, Source | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | 12783 | -0.145 | -0.4220 | Yes |
| 26 | GRN | GRN Entrez, Source | granulin | 12971 | -0.181 | -0.3657 | Yes |
| 27 | PRKRA | PRKRA Entrez, Source | protein kinase, interferon-inducible double stranded RNA dependent activator | 13016 | -0.194 | -0.2936 | Yes |
| 28 | ID3 | ID3 Entrez, Source | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | 13220 | -0.314 | -0.1866 | Yes |
| 29 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 13313 | -0.503 | 0.0022 | Yes |