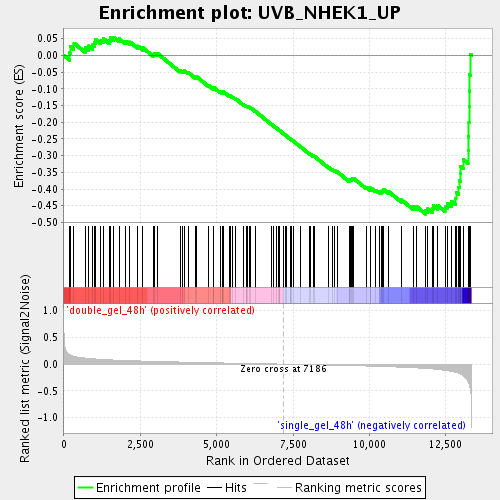
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | single_gel_48h |
| GeneSet | UVB_NHEK1_UP |
| Enrichment Score (ES) | -0.474913 |
| Normalized Enrichment Score (NES) | -1.7531452 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.049688637 |
| FWER p-Value | 0.998 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HMGCS2 | HMGCS2 Entrez, Source | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 (mitochondrial) | 190 | 0.175 | 0.0075 | No |
| 2 | MYF6 | MYF6 Entrez, Source | myogenic factor 6 (herculin) | 205 | 0.172 | 0.0279 | No |
| 3 | ATF3 | ATF3 Entrez, Source | activating transcription factor 3 | 329 | 0.145 | 0.0367 | No |
| 4 | HIST2H2AA3 | HIST2H2AA3 Entrez, Source | histone cluster 2, H2aa3 | 699 | 0.112 | 0.0228 | No |
| 5 | CDH16 | CDH16 Entrez, Source | cadherin 16, KSP-cadherin | 804 | 0.107 | 0.0283 | No |
| 6 | CEACAM1 | CEACAM1 Entrez, Source | carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) | 932 | 0.101 | 0.0313 | No |
| 7 | ITPR1 | ITPR1 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 1 | 1000 | 0.098 | 0.0384 | No |
| 8 | TUBB3 | TUBB3 Entrez, Source | tubulin, beta 3 | 1042 | 0.096 | 0.0472 | No |
| 9 | GAD2 | GAD2 Entrez, Source | glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) | 1212 | 0.089 | 0.0456 | No |
| 10 | IER2 | IER2 Entrez, Source | immediate early response 2 | 1296 | 0.086 | 0.0501 | No |
| 11 | TITF1 | TITF1 Entrez, Source | thyroid transcription factor 1 | 1498 | 0.079 | 0.0448 | No |
| 12 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 1528 | 0.079 | 0.0524 | No |
| 13 | SPINK1 | SPINK1 Entrez, Source | serine peptidase inhibitor, Kazal type 1 | 1631 | 0.076 | 0.0542 | No |
| 14 | GAST | GAST Entrez, Source | gastrin | 1807 | 0.072 | 0.0500 | No |
| 15 | NDUFA1 | NDUFA1 Entrez, Source | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1, 7.5kDa | 2019 | 0.068 | 0.0425 | No |
| 16 | TOB1 | TOB1 Entrez, Source | transducer of ERBB2, 1 | 2158 | 0.065 | 0.0401 | No |
| 17 | BTG1 | BTG1 Entrez, Source | B-cell translocation gene 1, anti-proliferative | 2418 | 0.060 | 0.0281 | No |
| 18 | BCL2L11 | BCL2L11 Entrez, Source | BCL2-like 11 (apoptosis facilitator) | 2584 | 0.057 | 0.0227 | No |
| 19 | PHLDA1 | PHLDA1 Entrez, Source | pleckstrin homology-like domain, family A, member 1 | 2920 | 0.051 | 0.0038 | No |
| 20 | S100A9 | S100A9 Entrez, Source | S100 calcium binding protein A9 | 2964 | 0.051 | 0.0069 | No |
| 21 | PPIF | PPIF Entrez, Source | peptidylprolyl isomerase F (cyclophilin F) | 3047 | 0.049 | 0.0069 | No |
| 22 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 3799 | 0.039 | -0.0450 | No |
| 23 | CRYBA4 | CRYBA4 Entrez, Source | crystallin, beta A4 | 3888 | 0.038 | -0.0469 | No |
| 24 | PIK3R3 | PIK3R3 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 3 (p55, gamma) | 3943 | 0.037 | -0.0463 | No |
| 25 | KYNU | KYNU Entrez, Source | kynureninase (L-kynurenine hydrolase) | 4061 | 0.035 | -0.0507 | No |
| 26 | CCK | CCK Entrez, Source | cholecystokinin | 4310 | 0.032 | -0.0654 | No |
| 27 | BCL2 | BCL2 Entrez, Source | B-cell CLL/lymphoma 2 | 4328 | 0.032 | -0.0627 | No |
| 28 | BRD2 | BRD2 Entrez, Source | bromodomain containing 2 | 4720 | 0.027 | -0.0888 | No |
| 29 | JUND | JUND Entrez, Source | jun D proto-oncogene | 4884 | 0.025 | -0.0979 | No |
| 30 | TAGLN3 | TAGLN3 Entrez, Source | transgelin 3 | 4905 | 0.025 | -0.0963 | No |
| 31 | DUSP1 | DUSP1 Entrez, Source | dual specificity phosphatase 1 | 5117 | 0.022 | -0.1095 | No |
| 32 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 5178 | 0.022 | -0.1113 | No |
| 33 | ATP5I | ATP5I Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit E | 5180 | 0.022 | -0.1087 | No |
| 34 | HTR4 | HTR4 Entrez, Source | 5-hydroxytryptamine (serotonin) receptor 4 | 5222 | 0.021 | -0.1091 | No |
| 35 | MAP1LC3B | MAP1LC3B Entrez, Source | microtubule-associated protein 1 light chain 3 beta | 5425 | 0.019 | -0.1220 | No |
| 36 | TRIB1 | TRIB1 Entrez, Source | tribbles homolog 1 (Drosophila) | 5445 | 0.019 | -0.1211 | No |
| 37 | PPBP | PPBP Entrez, Source | pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) | 5528 | 0.018 | -0.1250 | No |
| 38 | TACSTD2 | TACSTD2 Entrez, Source | tumor-associated calcium signal transducer 2 | 5622 | 0.017 | -0.1300 | No |
| 39 | BAMBI | BAMBI Entrez, Source | BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) | 5874 | 0.014 | -0.1471 | No |
| 40 | COX7A2 | COX7A2 Entrez, Source | cytochrome c oxidase subunit VIIa polypeptide 2 (liver) | 5968 | 0.013 | -0.1525 | No |
| 41 | PAX8 | PAX8 Entrez, Source | paired box gene 8 | 5982 | 0.013 | -0.1519 | No |
| 42 | CCNA1 | CCNA1 Entrez, Source | cyclin A1 | 6019 | 0.013 | -0.1531 | No |
| 43 | NR1D1 | NR1D1 Entrez, Source | nuclear receptor subfamily 1, group D, member 1 | 6058 | 0.012 | -0.1544 | No |
| 44 | EIF3S4 | EIF3S4 Entrez, Source | eukaryotic translation initiation factor 3, subunit 4 delta, 44kDa | 6121 | 0.012 | -0.1576 | No |
| 45 | RPS2 | RPS2 Entrez, Source | ribosomal protein S2 | 6263 | 0.010 | -0.1670 | No |
| 46 | NR4A2 | NR4A2 Entrez, Source | nuclear receptor subfamily 4, group A, member 2 | 6791 | 0.004 | -0.2063 | No |
| 47 | IL6 | IL6 Entrez, Source | interleukin 6 (interferon, beta 2) | 6864 | 0.004 | -0.2113 | No |
| 48 | RPS21 | RPS21 Entrez, Source | ribosomal protein S21 | 6942 | 0.003 | -0.2168 | No |
| 49 | EEF1A1 | EEF1A1 Entrez, Source | eukaryotic translation elongation factor 1 alpha 1 | 7012 | 0.002 | -0.2218 | No |
| 50 | RAP1A | RAP1A Entrez, Source | RAP1A, member of RAS oncogene family | 7041 | 0.002 | -0.2237 | No |
| 51 | TGFB1 | TGFB1 Entrez, Source | transforming growth factor, beta 1 (Camurati-Engelmann disease) | 7053 | 0.001 | -0.2244 | No |
| 52 | RPS17 | RPS17 Entrez, Source | ribosomal protein S17 | 7191 | -0.000 | -0.2347 | No |
| 53 | COL11A1 | COL11A1 Entrez, Source | collagen, type XI, alpha 1 | 7238 | -0.001 | -0.2381 | No |
| 54 | RPS23 | RPS23 Entrez, Source | ribosomal protein S23 | 7280 | -0.001 | -0.2410 | No |
| 55 | RPS27 | RPS27 Entrez, Source | ribosomal protein S27 (metallopanstimulin 1) | 7399 | -0.003 | -0.2496 | No |
| 56 | APOE | APOE Entrez, Source | apolipoprotein E | 7411 | -0.003 | -0.2501 | No |
| 57 | RPL31 | RPL31 Entrez, Source | ribosomal protein L31 | 7458 | -0.003 | -0.2532 | No |
| 58 | HCRT | HCRT Entrez, Source | hypocretin (orexin) neuropeptide precursor | 7499 | -0.004 | -0.2558 | No |
| 59 | RPL37 | RPL37 Entrez, Source | ribosomal protein L37 | 7734 | -0.006 | -0.2727 | No |
| 60 | SCG5 | SCG5 Entrez, Source | secretogranin V (7B2 protein) | 8047 | -0.010 | -0.2950 | No |
| 61 | UQCRB | UQCRB Entrez, Source | ubiquinol-cytochrome c reductase binding protein | 8079 | -0.010 | -0.2960 | No |
| 62 | ARHGDIA | ARHGDIA Entrez, Source | Rho GDP dissociation inhibitor (GDI) alpha | 8158 | -0.011 | -0.3005 | No |
| 63 | CHRNB4 | CHRNB4 Entrez, Source | cholinergic receptor, nicotinic, beta 4 | 8203 | -0.012 | -0.3024 | No |
| 64 | PPP2R1A | PPP2R1A Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), alpha isoform | 8667 | -0.018 | -0.3352 | No |
| 65 | MAOB | MAOB Entrez, Source | monoamine oxidase B | 8794 | -0.019 | -0.3423 | No |
| 66 | CLTB | CLTB Entrez, Source | clathrin, light chain (Lcb) | 8851 | -0.020 | -0.3440 | No |
| 67 | CXCR3 | CXCR3 Entrez, Source | chemokine (C-X-C motif) receptor 3 | 8942 | -0.021 | -0.3482 | No |
| 68 | PGK1 | PGK1 Entrez, Source | phosphoglycerate kinase 1 | 9333 | -0.026 | -0.3744 | No |
| 69 | STXBP2 | STXBP2 Entrez, Source | syntaxin binding protein 2 | 9357 | -0.027 | -0.3728 | No |
| 70 | ID2 | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 9384 | -0.027 | -0.3713 | No |
| 71 | RALY | RALY Entrez, Source | RNA binding protein, autoantigenic (hnRNP-associated with lethal yellow homolog (mouse)) | 9427 | -0.028 | -0.3710 | No |
| 72 | GUK1 | GUK1 Entrez, Source | guanylate kinase 1 | 9438 | -0.028 | -0.3683 | No |
| 73 | TOM1 | TOM1 Entrez, Source | target of myb1 (chicken) | 9478 | -0.029 | -0.3676 | No |
| 74 | MMP10 | MMP10 Entrez, Source | matrix metallopeptidase 10 (stromelysin 2) | 9894 | -0.035 | -0.3947 | No |
| 75 | PSMD8 | PSMD8 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 | 10029 | -0.037 | -0.4001 | No |
| 76 | YKT6 | YKT6 Entrez, Source | YKT6 v-SNARE homolog (S. cerevisiae) | 10045 | -0.038 | -0.3966 | No |
| 77 | GPI | GPI Entrez, Source | glucose phosphate isomerase | 10196 | -0.040 | -0.4029 | No |
| 78 | PFN1 | PFN1 Entrez, Source | profilin 1 | 10319 | -0.042 | -0.4070 | No |
| 79 | SURF1 | SURF1 Entrez, Source | surfeit 1 | 10391 | -0.043 | -0.4069 | No |
| 80 | IER3 | IER3 Entrez, Source | immediate early response 3 | 10439 | -0.044 | -0.4050 | No |
| 81 | HSP90AB1 | HSP90AB1 Entrez, Source | heat shock protein 90kDa alpha (cytosolic), class B member 1 | 10450 | -0.044 | -0.4002 | No |
| 82 | GADD45B | GADD45B Entrez, Source | growth arrest and DNA-damage-inducible, beta | 10614 | -0.047 | -0.4066 | No |
| 83 | MNAT1 | MNAT1 Entrez, Source | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 11046 | -0.057 | -0.4321 | No |
| 84 | IGFBP6 | IGFBP6 Entrez, Source | insulin-like growth factor binding protein 6 | 11430 | -0.066 | -0.4528 | No |
| 85 | NEU1 | NEU1 Entrez, Source | sialidase 1 (lysosomal sialidase) | 11548 | -0.070 | -0.4529 | No |
| 86 | H2AFX | H2AFX Entrez, Source | H2A histone family, member X | 11841 | -0.081 | -0.4648 | Yes |
| 87 | PCTK1 | PCTK1 Entrez, Source | PCTAIRE protein kinase 1 | 11904 | -0.084 | -0.4591 | Yes |
| 88 | DSCR1L1 | DSCR1L1 Entrez, Source | Down syndrome critical region gene 1-like 1 | 12052 | -0.090 | -0.4590 | Yes |
| 89 | CAPNS1 | CAPNS1 Entrez, Source | calpain, small subunit 1 | 12082 | -0.091 | -0.4498 | Yes |
| 90 | DYNLL1 | DYNLL1 Entrez, Source | dynein, light chain, LC8-type 1 | 12221 | -0.098 | -0.4480 | Yes |
| 91 | SGK | SGK Entrez, Source | serum/glucocorticoid regulated kinase | 12473 | -0.116 | -0.4525 | Yes |
| 92 | CYR61 | CYR61 Entrez, Source | cysteine-rich, angiogenic inducer, 61 | 12557 | -0.122 | -0.4436 | Yes |
| 93 | ANXA2 | ANXA2 Entrez, Source | annexin A2 | 12686 | -0.133 | -0.4366 | Yes |
| 94 | TSC22D1 | TSC22D1 Entrez, Source | TSC22 domain family, member 1 | 12825 | -0.152 | -0.4281 | Yes |
| 95 | CSRP1 | CSRP1 Entrez, Source | cysteine and glycine-rich protein 1 | 12832 | -0.153 | -0.4094 | Yes |
| 96 | CST6 | CST6 Entrez, Source | cystatin E/M | 12920 | -0.170 | -0.3948 | Yes |
| 97 | CYBA | CYBA Entrez, Source | cytochrome b-245, alpha polypeptide | 12938 | -0.173 | -0.3744 | Yes |
| 98 | CLDN4 | CLDN4 Entrez, Source | claudin 4 | 12969 | -0.181 | -0.3542 | Yes |
| 99 | HBEGF | HBEGF Entrez, Source | heparin-binding EGF-like growth factor | 12974 | -0.182 | -0.3317 | Yes |
| 100 | CHRNB1 | CHRNB1 Entrez, Source | cholinergic receptor, nicotinic, beta 1 (muscle) | 13070 | -0.218 | -0.3117 | Yes |
| 101 | CITED2 | CITED2 Entrez, Source | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | 13226 | -0.322 | -0.2832 | Yes |
| 102 | PDLIM5 | PDLIM5 Entrez, Source | PDZ and LIM domain 5 | 13230 | -0.326 | -0.2428 | Yes |
| 103 | CXCL2 | CXCL2 Entrez, Source | chemokine (C-X-C motif) ligand 2 | 13254 | -0.355 | -0.2002 | Yes |
| 104 | IL1RL1 | IL1RL1 Entrez, Source | interleukin 1 receptor-like 1 | 13263 | -0.378 | -0.1537 | Yes |
| 105 | CCL20 | CCL20 Entrez, Source | chemokine (C-C motif) ligand 20 | 13267 | -0.380 | -0.1064 | Yes |
| 106 | ADM | ADM Entrez, Source | adrenomedullin | 13274 | -0.393 | -0.0577 | Yes |
| 107 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 13313 | -0.503 | 0.0022 | Yes |